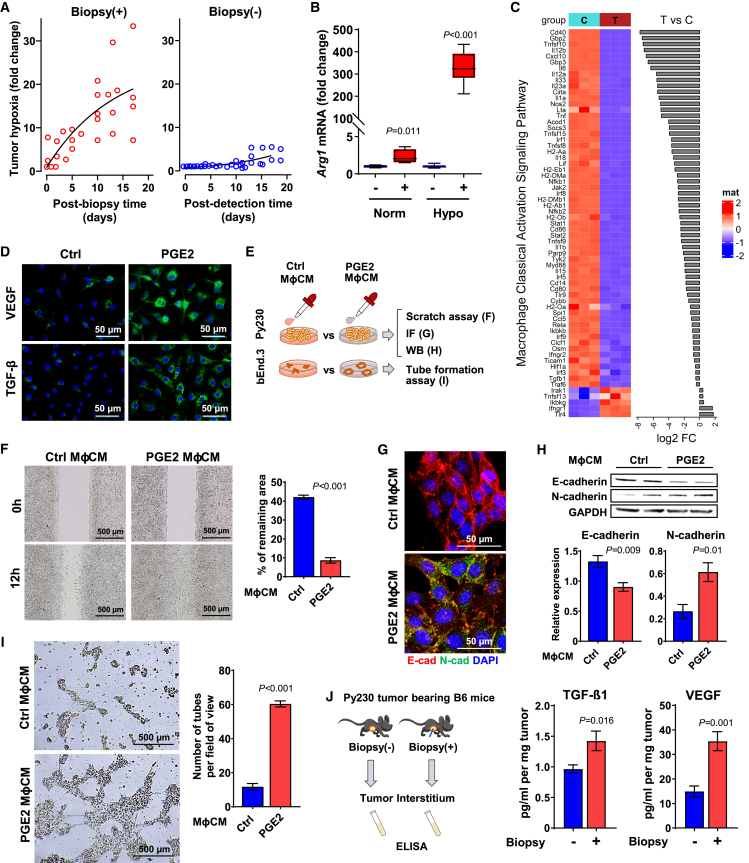
Figure 5.
PGE2 promotes M2 polarization under the hypoxic condition
(A) Tumor hypoxia was monitored longitudinally at indicated times from biopsied Py230 tumors (n = 5) or time-matched unbiopsied tumors (n = 6) using an IVIS in vivo imaging system and HypoxiSense 680 fluorescent probe. Time 0 indicates before biopsy. Regions of interest were quantified using Living Image software and are expressed in the graph as mean radiant efficiency over time. Data were analyzed in a linear mixed-effects model with random effects of mice and fixed effects of biopsy and time after biopsy.
(B) RT-qPCR analysis of Arg1 mRNA expression in the primary Mφ treated with or without PGE2 under normoxia (n = 4) or hypoxia (n = 8) and normalized by Gapdh. The data are shown as standard boxplots; p values were calculated using Student’s t test.
(C) Transcript expression analysis of IPA-selected classically activated Mφ (M1φ) in B6 control Mφ vs. PGE2-treated Mφ for 24 h under hypoxia (n = 3).
(D) Immunofluorescence (IF) images of VEGF and TGF-β1 of Mφ treated with or without PGE2 under hypoxia (n = 4).
(E) Schematic diagram of experimental flow of in vitro assays. Py230 and bEnd.3 cells were treated with conditioned medium (CM) collected from Mφ treated with PGE2.
(F) Wound-healing assay graph depicts the remaining areas after 12 h incubation, relative to untreated control (n = 9). p values were calculated using Student’s t test. The data are shown as mean ± SEM.
(G) IF images indicate Py230 cells treated with CM for 16 h (n = 4).
(H) Expression of E- and N-cadherin normalized by GAPDH (n = 5) in Py230 cells treated with MφCM for 16 h analyzed by western blot were compared using Student’s t test. The data are shown as mean ± SEM.
(I) Tube-formation assay of bEnd.3 cells treated with MφCM for 8 h (n = 3) were compared by Student’s t test. The data are shown as mean ± SEM.
(J) ELISA assays for TGF-β1 and VEGF-A of interstitial fluids collected from biopsied or unbiopsied Py230 tumors (n = 4) were calculated using Student’s t test. The data are shown as mean ± SEM.