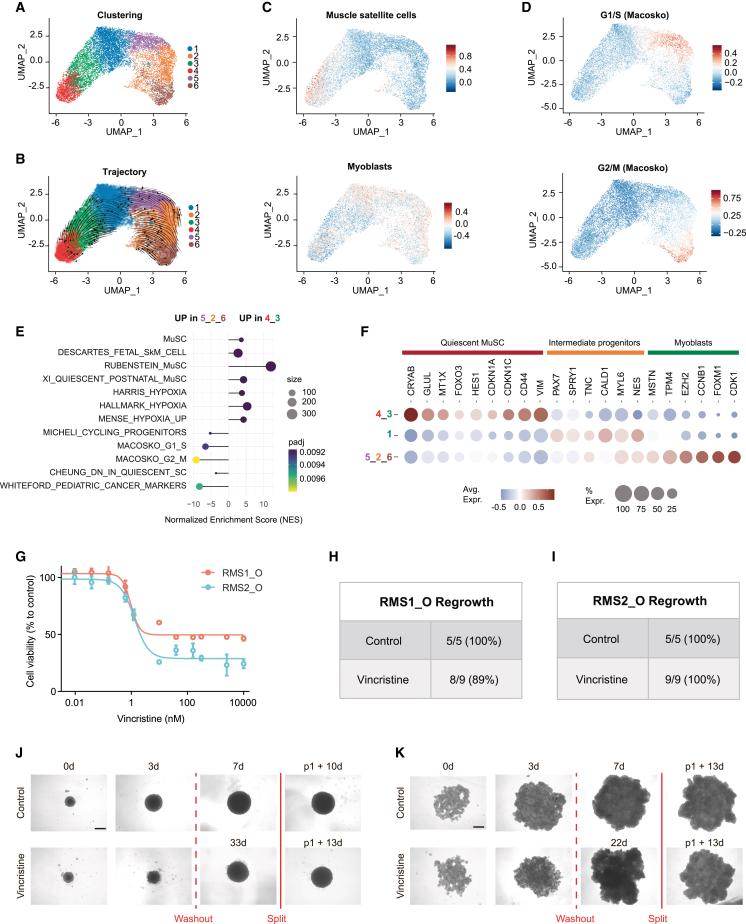
Figure 2.
FNRMS-derived organoids preserve functional intra-tumor heterogeneity
(A and B) scRNA-seq UMAP of RMS1_O showing cluster identities (A) and unsupervised trajectory inference analysis using scVelo (B). Data from 2 biological replicates at passages P13 and P14.
(C) Module scores of quiescent muscle satellite cell (top) and myoblast (bottom) expression programs displayed on scRNA-seq UMAP of RMS1_O.
(D) Module scores of cycling progenitors and G1/S (top) and G2/M (bottom) cell-cycle phases.
(E) Functional enrichment between quiescent muscle satellite-like cells (clusters 4-3) and myoblast proliferative cells (clusters 5-2-6) (see STAR Methods). Dots are colored according to their adjusted statistical probabilities with a yellow (lower significance) to blue (higher significance) gradient and sized by the count number of genes matching the biological process. MuSC, muscle satellite cells; SkM, skeletal muscle.
(F) Dot plot representing gene expressions of specific myogenic differentiation markers between cluster groups. Dots are sized according to the percentage of cells in each cluster group that express the gene (transcript level > 0) and color coded by average gene expression levels across cells.
(G–K) Efficiency of vincristine on RMS_Os.
(G) Vincristine dose-response curves performed on RMS_Os. Viability is expressed as a percentage of the value in untreated cells (CellTiter-Glo). Means ± SD are represented (n = 3).
(H–K) RMS_Os were treated or not with vincristine (5 nM) for 3 days. Treatments were then stopped, and regrowth of structures was evaluated within 50 days in each case.
(H and I) RMS1_O (H) and RMS2_O (I) regrowth rate within 30 days after treatment washout.
(J and K) Representative bright-field images of RMS1_O (J) and RMS2_O (K) at t = 0 and 3 days, with or without (vehicle) vincristine treatment. When regrowth of tumoroids was observed and tumoroids reached a size of 4–6 × 105 μm2, they were split to ensure that their renewal properties were preserved. WO, washout. Scale bar: 400 μm.