Abstract
It is long observed that females tend to live longer than males in nearly every country. However, the underlying mechanism remains elusive. In this study, we discovered that genetic associations with longevity are on average stronger in females than in males through bio-demographic analyses of genome-wide association studies (GWAS) dataset of 2178 centenarians and 2299 middle-age controls of Chinese Longitudinal Healthy Longevity Study (CLHLS). This discovery is replicated across North and South regions of China, and is further confirmed by North-South discovery/replication analyses of different and independent datasets of Chinese healthy aging candidate genes with CLHLS participants who are not in CLHLS GWAS, including 2972 centenarians and 1992 middle-age controls. Our polygenic risk score analyses of eight exclusive groups of sex-specific genes, analyses of sex-specific and not-sex-specific individual genes, and Genome-wide Complex Trait Analysis using all SNPs all reconfirm that genetic associations with longevity are on average stronger in females than in males. Our discovery/replication analyses are based on genetic datasets of in total 5150 centenarians and compatible middle-age controls, which comprises the worldwide largest sample of centenarians. The present study's findings may partially explain the well-known male-female health-survival paradox and suggest that genetic variants may be associated with different reactions between males and females to the same vaccine, drug treatment and/or nutritional intervention. Thus, our findings provide evidence to steer away from traditional view that “one-size-fits-all” for clinical interventions, and to consider sex differences for improving healthcare efficiency. We suggest future investigations focusing on effects of interactions between sex-specific genetic variants and environment on longevity as well as biological function.
Keywords: Longevity, Male-female health-survival paradox, sex differences, Polygenic risk score analysis, Centenarians, Precise healthcare for males and females
1. Introduction
In general, females live longer and are less susceptible to mortality from infectious and non-communicable diseases compared to males [1,2]. The increased life expectancy of females as a phenomenon became apparent when data for people born in the late 19th century became available [3]. This data also suggested that the improved medical and social status of females during the last century may have benefitted females more than males with respect to longevity. Additional studies showed that males have had significantly higher mortality rates than females during famines and other natural disasters, worldwide [[4], [5], [6]]. During the European summer heat wave of 2003, among persons aged 65 or older, age-specific mortality rates of males were twice as high as those of females [7]. In countries where the data are available (China, South Korea, Italy, Spain, Denmark, Mexico, Portugal, Greece, Norway, England, Philippines, Slovenia, Czech, South Korea, Italy, and Spain), age-specific fatality rates have been substantially higher among male COVID-19 patients than among their female counterparts, and the sex gap quickly increase with age after age 50 [[8], [9], [10], [11], [12]].
In general, the socioeconomic status, including education, income and occupation, as well as health status, as measured by functional capacities in activities of daily living, cognition, and physical performance, are substantially worse in females than in males in China [13] and elsewhere [14,15]. The phenomenon that females on average live significantly longer than males in spite of having poorer health and lower socioeconomic status is known as the male-female health-survival paradox [1].
The scope of the influence of genetics on this paradox is not known. Previously published genome-wide association studies (GWAS) of longevity adjusted for sex as a covariate, identified sex-independent genetic risk factors, such as APOE (Apolipoprotein E), FOXO1A (Forkhead Box O1A) and IL6 (Interleukin 6) [[16], [17], [18], [19]], but did not investigate the sex-specific genesgenes [20]. Other studies have revealed significant sex-specific associations of genetic variants with different diseases and health outcomes [21]. The functional EXO1 (Exonuclease 1) gene promoter variant is associated with increased life expectancy in female centenarians only [22]. FOXO3 is the second most-replicated gene with longevity [[23], [24], [25], [26]], and several studies conducted using Western populations showed that it is a sex-specific longevity gene that favors males more than females [23,24]. However, this male-specific effect may not be universally valid since a recent study showed that FOXO3 was protective in females and not males; the same study showed SIRT1 (Sirtuin 1) was associated with longevity in males [26].
EXO1, FOXO3 and SIRT1 are single genes and don't reveal whether polygenetic associations with longevity are on average statistically more significant in males or females, which is jointly determined by small effects of numerous genes. Based on sex-specific GWAS on the Chinese Longitudinal Healthy Longevity Study (CLHLS) datasets, four groups of sex-specific genes were found to be associated with longevity [20]. However, the study did not investigate the potential statistically more significant effect of polygenetic associations with longevity in females compared to males and vice versa.
Although research on sex differences in mortality and the socioeconomic and behavioral determinants have proliferated recently [15], as yet no study has addressed whether polygenetic associations with longevity are on average more significant in one sex or the other. The present study aims to address this important scientific question to better understand the male-female health-survival paradox.
2. Results
2.1. Identification of eight groups of sex-specific genes jointly associated with longevity
We have identified eight groups of genes that are jointly associated with longevity at different significance levels. We identified these eight groups using datasets of CLHLS GWAS and CLHLS healthy aging candidate genes, and they are listed in SM-Table 3a-3b. Table 1 summarizes the criteria for identifying each of these sex-specific groups: the set of sex-specific longevity genes were grouped by jointly-significant association (P < 10−8) with longevity in one sex but in the other sex (P > 0.05) based on polygenic risk score (PRS) analysis [20]. By using the PRSice software, the PT values were implemented for searching an ideal threshold PT to find the best-fit [27]. Meanwhile, the approach used to analyze every individual sex-specific longevity associated gene at a pre-defined significant P value in one sex but not the other (PT > 0.05) was described in SM4.
Table 1.
Comparisons of the average probabilities of longevity between males and females with various standardized PRS summarizing each of the eight groups of sex-specific genes jointly associated with longevity at different significance levels.
| Average probability of longevity |
Ave. prob. of longevity in females higher than in males |
P value of sex difference |
|||
|---|---|---|---|---|---|
| Females | Males | Diff. | % of diff. | ||
| A. Analyses based on the CLHLS GWAS datasets | |||||
| (I) Sex-specific top genes jointly associated with longevity | |||||
| (1) 11 male-specific top genes jointly associated with longevity (each of them has P < 10−5 in males, but P > 0.16 in females) | 0.513 | 0.420 | 0.093 | 22.15 % | 8.4 × 10−09 |
| (2) 11 female-specific top genes jointly associated with longevity (each of them has P < 10−5 in females, but P > 0.12 in males) | 0.514 | 0.423 | 0.090 | 21.34 % | 3.8 × 10−08 |
| (II) Sex-specific strong genes jointly associated with longevity | |||||
| (3) 35 male-specific strong genes jointly associated with longevity (each of them has 10−5≤P < 10−4 in males, but P > 0.25 in females) | 0.513 | 0.420 | 0.093 | 22.10 % | 1.9 × 10−08 |
| (4) 25 female-specific strong genes jointly associated with longevity (each of them has 10−5≤P < 10−4 in females, but P > 0.36 in males) | 0.516 | 0.423 | 0.093 | 22.06 % | 7.8 × 10−09 |
| (III) Sex-specific moderate genes jointly associated with longevity | |||||
| (5) 191 male-specific moderate genes jointly associated with longevity (each of them has 10−4≤P < 10−3 in males, but P > 0.75 in females) | 0.514 | 0.415 | 0.099 | 23.95 % | 2.0 × 10−09 |
| (6) 311 female-specific moderate genes jointly associated with longevity (each of them has 10−4≤P < 10−3 in females, but P > 0.70 in males) | 0.515 | 0.422 | 0.093 | 21.93 % | 1.5 × 10−08 |
| B. Analyses based on the CLHLS healthy aging candidate genes datasets | |||||
| (IV) Sex-specific genes jointly associated with longevity | |||||
| (7) 30 Male-specific jointly associated with longevity (each of them has P < 0.05 in males, but P > 0.10 in females) | 0.701 | 0.570 | 0.130 | 22.88 % | 3.0 × 10−47 |
| (8) 74 female-specific genes jointly associated with longevity (each of them has P < 0.05 in females, but P > 0.10 in males) | 0.689 | 0.540 | 0.149 | 27.53 % | 1.1 × 10−45 |
Using these analytic approaches, we found that each of the eight groups of sex-specific genes listed in SM-Table 3a-3b are jointly and significantly associated with longevity in one sex (P = 8.7 × 10−165∼1.5 × 10−37), but not jointly significant in the other sex (P > 0.05), while PRS-sex interaction effects are highly significant (P = 5.2 × 10−106∼4.4 × 10−15) (SM-Table 5). To simplify the presentation of the eight groups of sex-specific genes, we only indicate individual P values for males and females of each group of genes throughout the text, and do not indicate the group's joint-significance P values which are presented in SM-Table 5.
2.2. Comparisons of the average probabilities of longevity between males and females
Table 1 also presents the average probabilities of longevity conferred by each of the eight groups of longevity genes (formula (4)). The average probability of longevity is the likelihood that an individual with a given PRS value is a centenarian, as estimated by the logistic regression (Methods section M3). Table 1 Section A summarizes the results of analysis of the CLHLS GWAS datasets, where the average probabilities for females were substantially higher than in males by a margin of 21.3 %–24.0 % for all significance thresholds (see panels (I)-(III) in Table 1, Section A). The analysis summarized in Table 1 Section B utilized the CLHLS healthy aging candidate gene datasets, and confirmed this result, namely, that the average probabilities of longevity are substantially higher in females than in males in each group of the sex-specific longevity genes.
2.3. The female to male ratio of relative benefits associated with the sex-specific longevity genes
To quantify the sex differences in the benefits due to genetic associations with longevity, we estimate and compare female-to-male ratios of relative benefits (compared with the other sex) associated with male-specific genes of longevity and female-specific genes of longevity, respectively, employing a novel bio-demographic method described in Methods section M4, that is based on logistic regression analyses on standardized PRS. These results are summarized in Fig. 1a and b, Fig. 2a and b, Fig. 3a and b, Fig. 4a and b.
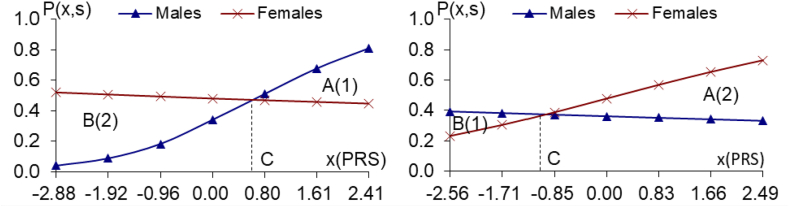
Fig. 1a and b.
The PRS analyses on the 11/11 male/female-specific top genes of longevity (P < 10−5 in one sex, but P > 0.12 in another sex), using the CLHLS GWAS datasets Fig. 1a. 11 male-specific top genes of longevity Fig. 1b. 11 female-specific top genes of longevity, (P < 10−5 in males, but P > 0.16 in females)(P < 10-5 in females, but P > 0.12 in males)
Notes: (1) P(x,s) in the vertical axis denotes the probability of longevity among persons with sex s (s = 1, male; s = 2, female) and the standardized polygenic risk score (PRS) value x which summarizes propensity to longevity of a group of sex-specific genes; P(x,s) is estimated based on the logistic regression model described in the Method section M3; (2) x in the horizontal axis represent the PRS values which is estimated by the procedure described in the Methods section M2; C is the PRS value at which the probabilities of longevity in males and females are equal to each other (i.e., the male and female curves crossover at x(PRS) C); (3) The A(1), B(2), FM1, A(2), B(1) and FM2 are estimated using the method described in the Methods section M4.
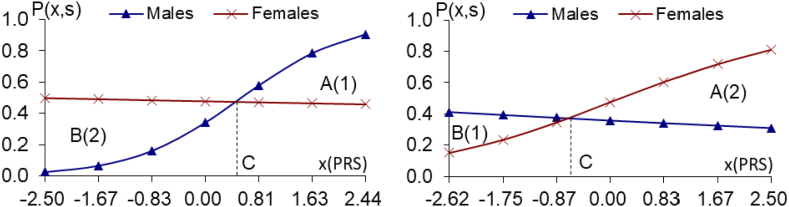
Fig. 2a and b.
The PRS analyses on the 35/25 male/female-specific strong genes of longevity (10−5≤P < 10−4 in one sex, but P > 0.25 in another sex), using the CLHLS GWAS datasets, Fig. 2a. 35 male-specific strong genes of longevity Fig. 2b. 25 female-specific strong genes of longevity (10−5≤P < 10−4 in males, but P > 0.25 in females), (10−5≤P < 10-4 in females, but P > 0.36 in males) Notes: the same as the notes in Fig. 1a and b.
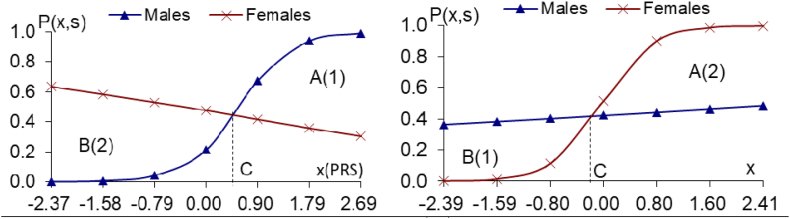
Fig. 3a and b.
The PRS analyses on the 191/311 male/female-specific moderate genes of longevity (10−4≤P < 10−3 in one sex, but P > 0.70 in another sex), using the CLHLS GWAS datasets Fig. 3a.191 male-specific moder. genes of longevity Fig. 3b. 311 female-specific moder. genes of longevity, (10−4≤P < 10−3 in males, but P > 0.75 in females), (10−4≤P < 10−3 in females, but P > 0.70 in males), Notes: the same as the notes in Fig. 1a and b.
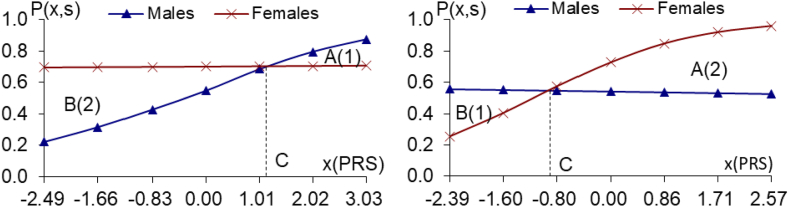
Fig. 4a and b.
The PRS analyses on the 30/74 male/female-specific genes of longevity (P < 9.0 × 10−3 in one sex, but P > 0.10 in another sex), using the CLHLS healthy aging candidate genes datasets, Fig. 4a. 30 male-specific genes of longevity Fig. 4b. 74 female-specific genes of longevity (P < 9.0 × 10−3 in males, but P > 0.10 in females), (P < 8.0 × 10−3 in females, but P > 0.12 in males)Notes: the same as the notes in Fig. 1a and b.
2.3.1. The female to male ratio of relative benefits due to male-specific genes of longevity
As shown in Fig. 1a, the probabilities of longevity associated with the positive joint effects of higher PRS (i.e., PRS > C) of the 11 male-specific genes are greater for males than for females, but when PRS < C, the probabilities of longevity for females is greater than for males because the female curve in Fig. 1a is above the male curve.
Fig. 1a also indicates that the relative benefits attributed to the 11 male-specific top genes of longevity (P < 10−5) are higher for females than males. This assessment is based on the relative areas between the respective PRS curves on either side of the cross-over point C. The male relative benefit when PRS > C of these 11 male-specific top genes associated with longevity (P < 10−5) is 34.71, i.e., the triangle area A(1) in Fig. 1a. This is the sum of differences in probabilities of longevity between males and females when PRS > C. By contrast, relative benefits accrue to females when PRS < C of the 11 male-specific top longevity-associated genes because the sum of differences in probabilities of longevity between females and males, the triangle area B(2) in Fig. 1a, is 105.59. Consequently, the female-to-male ratio of relative benefits accounts for these 11 male-specific top genes of longevity is 3.04 (FM1 = B(2)/A(1) = 105.59/34.71), and the PRS-sex interaction effects are highly significant (P = 9.7 × 10−19), as described in Fig. 1a legend.
The same pattern holds true for the 35 male-specific strong genes (Fig. 2a) or the 191 male-specific moderate genes (Fig. 3a), namely that the relative benefits are all substantially higher in females than in males). The female-to-male ratios of relative benefits due to 35 male-specific strong genes of longevity (10−5≤P < 10−4, Fig. 2a) or 191 male-specific moderate genes of longevity (10−4≤P < 10−3, Fig. 3a) are all substantially larger than one, and the PRS-sex interaction effects are all highly significant (see Figs. 2a–3a legends).
2.3.2. The female to male ratio of relative benefits due to female-specific genes of longevity
The female relative benefits due to the positive joint effects of higher PRS (i.e. PRS > C) of the 11 female-specific top genes of longevity (P < 10−5) are 71.50 (area A(2) in Fig. 1b), which is the sum of differences in probabilities of longevity between females and males, all with PRS higher than C. On the other hand, when PRS < C, the male relative benefits of the 11 female-specific top genes of longevity are 13.21 (area B(1)). Thus, the female-to-male ratio of relative benefits due to the 11 female-specific top genes of longevity is 5.41 (FM2 = A(2)/B(1) = 71.50/13.21), and the PRS-sex interaction effects are highly significant (P = 9.1 × 10−8) (Fig. 1b legend). These results indicate that the relative benefits due to the 11 female-specific top genes of longevity (P < 10−5) are more apparent in females.
The estimates presented in Figs. 2b–3b, based on the CLHLS GWAS datasets, reveal the same pattern as in Fig. 1b, namely, the relative benefits due to the 25 female-specific strong genes of longevity (10−5≤P < 10−4) or the 311 female-specific moderate genes of longevity (10−4≤P < 10−3) are all substantially much higher in females than in males (Figs. 2b–3b legends).
2.4. Confirmation analyses using the CLHLS healthy aging candidate genes datasets
To further confirm the results based on the CLHLS GWAS datasets described above, we conducted additional South-North evaluation/replication PRS analyses on 30 male-specific and 74 female-specific genes of longevity (P < 9.0 × 10−3; Fig. 4a–b. These genes were identified and replicated individually, based on totally independent datasets of the CLHLS participants, which included 2972 centenarians and 1992 middle-age controls (SM3). The female to male ratios of relative benefits due to the 30 male-specific genes of longevity or 74 female-specific genes of longevity are all substantially larger than one, and the PRS-sex interaction effects are highly significant, confirming that the relative benefits due to sex-specific genes of longevity are much higher in females than in males.
2.5. Additional PRS analysis of randomly assigned female centenarian/control samples that have exactly the same size as that of male centenarians/controls
It is common in studies of centenarians to have more female than male participants. To test whether our findings are biased by this difference in sample sizes, we conducted additional analyses based on a sample constructed of equally sized groups stratified by sex: 564 male centenarians and 564 randomly selected female centenarians, and 773 male middle-age controls and 773 randomly selected female middle-age controls, using both the CLHLS GWAS and CLHLS healthy aging candidate genes datasets. The results of this analyses were consistent with our prior results, namely, genetic associations with longevity are on average stronger in females than in males (SM-Table 7 and SM-Figs. 1–4). This indicates that our current results are not affected by the larger sample size of female centenarians than their male counterparts. The higher death rate among males may increase the statistical power of the male centenarian sample [20].
2.6. Analyses of all genes individually associated with longevity in both sexes or in one sex only
The genetic contributions to longevity in males and females are not attributed only to groups of sex-specific genes jointly associated with longevity (as outlined in section 2.1), but genes that are individually associated with longevity in both sexes or in one sex only also contribute. Thus, to obtain a complete picture of whether genetic associations with longevity are stronger in males or females, we further conducted logistic regression analyses on all genes that are individually associated with longevity in both sexes or in one sex only, and we used P < 0.05 as the individually significant P threshold.
2.6.1. Genes individually associated with longevity in both sexes
Using the CLHLS GWAS and the CLHLS healthy aging candidate genes datasets, we identified three groups of the genes that are individually associated with longevity in both sexes at different significance levels (SM Tables 8a–8c). These groups differed by the stringency of the significance of association with longevity. One group, including 17 individual genes, had P < 10−4 in one sex and P < 0.05 in the other (SM Table 8a). The second group, composed of 92 genes, exhibited 10−4<P < 10−3 in one sex and P < 0.05 in the other (SM Table 8b). The third group, composed of 128 genes, exhibited P < 0.05 in both sexes (SM Table 8c). For each group, the PRS values favor females over males SM Fig. 5ã5c). The average probabilities of longevity of these three groups of genes are higher in females than in males by 23.4 %–32.9 %, and the sex differences are highly significant (Table 2 A, panel I).
Table 2.
Comparisons of the average probabilities of longevity between males and females with various standardized PRS summarizing each of the groups of genes individually associated with longevity in both sex or in one sex only.
| Average probability of longevity |
Ave. prob. of longevity in females higher than in males |
P value of sex difference |
|||
|---|---|---|---|---|---|
| Females | Males | Diff. | % of diff. | ||
| A. Analyses based on the CLHLS GWAS datasets | |||||
| (I) The genes individually associated with longevity in both sex | |||||
| 17 genes individually associated with longevity in both sex (P < 10−4 in one sex and P < 0.05 in other sex) | 0.424 | 0.319 | 0.105 | 32.88 % | 2.0 × 10−20 |
| 92 genes individually associated with longevity in both sex (10−4≤P < 10−3 in one sex and P < 0.05 in the other sex) | 0.468 | 0.379 | 0.089 | 23.41 % | 8.8 × 10−14 |
| (II) The genes individually associated with longevity in one sex only | |||||
| 190 genes individually associated with longevity in one sex only (P < 10−4 in one sex but P ≥ 0.05 in other sex) | 0.483 | 0.414 | 0.069 | 16.69 % | 0.0002 |
| 1367 genes individually associated with longevity in one sex only (10−4≤P < 10−3 in one sex but P ≥ 0.05 in the other sex) | 0.519 | 0.474 | 0.044 | 9.31 % | 0.095 |
| B. Analyses based on the CLHLS healthy aging candidate genes datasets | |||||
| (III) The genes individually associated with longevity in both sex | |||||
| 128 genes individually associated with longevity (P < 0.05 in both sex) | 0.690 | 0.555 | 0.135 | 24.32 % | 4.4 × 10−20 |
| (IV) The genes individually associated with longevity in one sex only | |||||
| 152 genes associated with longevity in one sex only (P < 0.05 in one sex but P ≥ 0.05 in the other sex) | 0.698 | 0.532 | 0.165 | 31.08 % | 5.2 × 10−172 |
Notes: (a) The estimates presented in this table are based on the logistic regressions including continuous standardized PRS, sex and (PRS x sex) interaction term (ref. Equations (2), (3) in the Methods section M3); (b) The sex-specific average probabilities of longevity among persons of sex s with various PRS values presented in this table are estimated using formula (4) in Methods section M3; (c) The P values of the sex differences presented in the last column of this table are estimated using the bootstrap method (the number of bootstrap replications is set to 2000) of the statistical software STATA 12.0.
2.6.2. Genes individually associated with longevity in one sex only
We also identified three groups of the genes that are individually associated with longevity in one sex only at different significance levels (SM Tables 8d–8f), using the CLHLS GWAS datasets and the CLHLS healthy aging candidate genes datasets. The probabilities of longevity of various PRS values for each of these groups of genes also showed that genetic associations with longevity are on average stronger in females than in males (SM Fig. 6a–c), Table 2, section A(II) and B (II) demonstrate that in females the average probabilities of longevity of the groups of genes individually associated with longevity in one sex only are higher than those in males by 9.3 %–31.1 %, and the sex differences are highly significant, with one exception (P = 0.095).
2.7. Sex-based pathways associated with longevity
Next, we used the FUMA tool to map this significant longevity associated genes to the KEGG and REACTOME databases [28]. (SM Fig. 7a, P < 0.05), and neuroactive ligand receptor interaction Notably, the male-specific genes were mainly enriched in the extracellular matrix-related pathways, including glycosaminoglycan biosynthesis-heparan sulfate, NABA ECM glycoproteins, and cell-cell junction organization (SM Fig. 7a, P < 0.05). The association of altered extracellular matrix with age-associated diseases, such as Alzheimer's disease, has been widely documented [29]. By contrast, the female-specific genes were mainly enriched in the pathway of neuroactive ligand receptor interaction (SM Fig. 7b, P < 0.05).
2.8. Analyses of genetic heritability of longevity in males and females using all SNPs and the genome-wide Complex Trait Analysis (GCTA) method
Using the GCTA method (section 2.5) [30], we estimated the genetic heritability of longevity (h2) for each sex, based on analyzing all SNPs of the CLHLS GWAS and healthy aging candidate genes datasets. The male and female h2 are 0.172 and 0.187 respectively based on the CLHLS GWAS dataset; the male and female h2 are 0.199 and 0.255 respectively based on the CLHLS healthy aging candidate genes dataset. Clearly, the genetic heritability of longevity is stronger in females than that in males. These results indicate that the GCTA analyses using all SNPs of the CLHLS GWAS datasets and healthy aging candidate genes datasets reconfirm that genetic effects on longevity are on average stronger in females than in males.
3. Discussion
Based on the statistical and bio-demographic analyses of CLHLS GWAS datasets (including 2178 centenarians and 2299 middle-age controls), we discovered that average probabilities of longevity, associated with each of the six exclusive groups of sex-specific genes at tiered significance levels, are all significantly higher in females than in males. Our bio-demographic analyses demonstrate that the female-to-male ratios of relative benefit due to each of the six exclusive groups of sex-specific genes associated with longevity are all substantially in favor of females, and the PRS-sex interaction effects are all highly significant. This discovery that females are genetically ‘advantaged’ with respect to polygenetic associations with longevity was replicated across different datasets of South and North regions of China. Additional analyses of both sex-specific and non-sex-specific genes confirmed that polygenetic associations with longevity are on average stronger in females more than males.
We further confirmed this conclusion using independent datasets of CLHLS healthy aging candidate genes (including 2972 centenarians and 1992 middle-age controls). Moreover, the GCTA analyses, using all SNPs of both the CLHLS GWAS datasets and healthy aging candidate genes datasets, respectively, add an additional level of confirmation, that genetic effects on longevity are on average stronger in females than in males. Interestingly, a recent study based on results from a large GWAS on human longevity (N ≈ 390,000) demonstrated that a higher genetic predisposition for longevity had a stronger association with behavioral phenotypes (such as education, smoking, body mass index (BMI) and depression) in females than in males [31], which is concordant with our findings.
Our results beg the question of why are genetic associations with longevity on average stronger in females than in males? The fact that females take much more care for childbearing and offspring than males may shed light on answering this question. Studies related to age-specific manifestation of genetic load suggest that fertility serves as the major factor of Darwinian natural selection for the accumulation of genetic mutation driving population survival and growth [32]. The grandmother hypothesis [33] proposed that postmenopausal longevity in human evolved from grandmothers' assistance with childcare, which prolonged females’ lifespan.
A study reported that female centenarians were four times more likely to have children in their forties than females lived only to age 73 [34]. Other studies (including analyses based on the CLHLS datasets) also found that females’ late childbearing after ages 35 or 40 is positively and significantly associated with longevity [35,36]. A study indicated that the longevity advantage of females over males may be a by-product of genetic evolution that maximizes the length of time during which females could bear and take care of children and contribute to human reproduction [37].
The reproductive function of females might serve as a driving force for positive selection on the human genome and the related physiological features, such as immune response and metabolism. During periods of stress such as starvation, females use available amino acids to create deposits in the liver to support reproduction; conversely males slow down anabolic pathways and reserve carbohydrate stores for eventual use by the musculature [38]. Sex differences in genetics also affect innate and adaptive immunity [39]. Various studies have reported a more progressive decline in immunity and dysregulated inflammatory response with increase of age in males than in females [40,41]. In the current study, our pathway analysis revealed neuronal system, glycosaminoglycan biosynthesis-heparan sulfate, NABA ECM glycoproteins, and cell-cell junction organization are male-specific pathways, and neuroactive ligand receptor interaction is the female-specific pathway. Interestingly, it is previously reported that reductions of heparan sulfate biosynthetic gene function increased lifespan in Drosophila parkin mutants [42].
The results of our study lead us to conclude that genetic associations with longevity are on average stronger in females than in males. This novel finding contributes to understanding the “male-female health-survival paradox”. We believe the genetic associations with longevity that are on average stronger in females than in males discovered in the present study are not driven by the fact that female centenarians and middle-age controls on average live longer than their male counterparts. This is because all GWAS of longevity (including present study) and our replication/reconfirmation analyses are not based on sex differences in lifespan among cases of centenarians and middle-aged controls. Instead, all male and female centenarians are counted as longevity “cases”, regardless of their lifespan differences, and all middle-aged male and female individuals are counted as “controls”, disregarding their lifespan differences. As explained in eAppendix section S1 of the article [19], all prior GWAS of longevity [[16], [17], [18]] and the present study investigate genetic associations with longevity by comparing the cross-sectional frequencies of carrying the genetic variants between centenarian cases and middle-aged controls, with no effects of the survival time with respect to each of the male and female centenarians and middle-age controls.
Our study has limitations. Similar to all other case/control association studies, including GWAS, this study could not empirically reveal the causalities of our findings, warranting further in-depth investigations. Moreover, we could not test the hypothesis that females' genetic advantage of longevity may be partially due to their XX chromosomes compared to males’ single X chromosome [43,44], because the CLHLS GWAS of longevity did not include SNPs on the X and Y chromosomes and mitochondria, due to genotyping technical reasons, which are the same as for other GWAS [16,17]. In addition, sex-specific longevity might also be influenced by other potential confounding factors, such as inherent gender inequality, hormonal differences, environmental exposures, and behavioral factors, which are not covered in this study. Despite these limitations, the new findings of this study warrant further investigation by interdisciplinary collaborations, such as validation using datasets from other countries, international meta-analysis with much larger sample sizes, and laboratory tests on biological functions.
Nevertheless, the findings of this study provide evidence supporting the notion that significant contributions of genetic factors to sex-biased lifespan and healthspan, and also might be helpful to develop prevention and treatment strategies for both male and female elderly patients with chronic diseases in this forthcoming era of precision medicine. Additionally, our study provides invaluable insight into further understanding of molecular mechanisms underlying sex differential aging related diseases as well as regulatory networks.
4. Conclusion
Our study of sex-specific GWAS and novel bio-demographic analyses have contributed to a better understanding of sex differences in genetic variants that may very likely lead to different reactions to the same vaccine, drug treatment and/or nutritional intervention, and also has potential application in studies of sex-specific differences in Alzheimer's disease [45,46]. Thus, steering away from the traditional view that “one-size-fits-all”, and considering sex differences may likely improve the healthcare efficiency [47]. We suggest that further investigations should focus on effects of interactions between sex-specific genetic variants and environmental factors on healthy aging and the biological mechanisms, which will substantially contribute to more effective precise healthcare for males and females.
Funding
This study is supported by the National Key R&D Program of China (2018YFC2000404; XYL, YZ), the National Natural Science Foundation of China (72061137004; XYL, YZ), and the United States NIA/NIH (P01AG031719, YZ, JWV; P30AG028716, AMO).
Role of funding source
The funding agencies provided financial support to the data and DNA samples collections and analyses, but they did not play any role in writing, interpreting the results and submission for consideration of publication of this manuscript. We are not paid by anyone to write this article. Yi Zeng, as the 1st corresponding author, has final responsibility for the decision to submit for publication.
Methods
M1. The genetic datasets analyzed in present study
We firstly analyze the CLHLS GWAS datasets, which include 5.6 million single nucleotide polymorphisms (SNPs) for each of 2178 centenarians and 2299 middle-age controls (ref. Supplementary Materials (SM) sections SM1∼SM2 and SM-Table 1). Secondly, to replicate the results, we analyzed independent datasets of CLHLS healthy aging candidate genes of 2972 centenarians and 1992 middle-age controls aged <65, who are CLHLS participants but not included in the CLHLS GWAS (SM-Table 2). Our healthy aging candidate genes datasets include 287,898 SNPs for each of the CLHLS participants. These 287,898 SNPs associated with longevity and chronic diseases were selected based on GWAS datasets of CLHLS and U.S. Health and Retirement Surveys as well as other published genetic databases. The present study is based on genetic datasets of in total 5150 centenarians and compatible middle-age controls, which has worldwide largest sample of centenarians so far, 3.9 times as large as worldwide second largest genetic datasets sample of 1320 centenarians [20]. Note that a wide variety of internationally published studies have confirmed that age reporting of Han Chinese centenarians is reasonably accurate [48,49].
To increase statistical rigor, we adopt a replication framework of South and North regions of China as discovery and evaluation/replication samples (SM-Table 1, Table 2), following most published case/control genetic studies that use Chinese nationwide datasets and based on analyses of principal components, genetics, anthropology, and linguistics, reported in the literature [50].
The Research Ethics Committees of Peking University and Duke University granted approval for the Protection of Human Subjects of CLHLS, including collections of the data and DNA samples as well as productions of the de-identified genotype and phenotype data used for present study. The survey respondents gave informed consent before participation. The uses of the genotype and phenotype data in this study are carried out in accordance with national and International legislative and institutional guidelines and regulations.
M2. The standardized polygenic risk score (PRS) analyses
It is impossible to address the present study's research question based solely on sex-specific single genes analysis, because each locus has a small effect and combinations of many genes contribute to the association with the phenotype [27,51]. Thus, we conducted analyses of PRS, which summarizes the joint effects of propensity for longevity of each of the groups of identified sex-specific genes, to explore whether the genetic associations with longevity are stronger in males or females.
Following conventional methodology, we constructed PRS of each of the identified groups of sex-specific genes for each of the centenarians and middle-age controls, using the odds ratios estimated from the discovery sub-dataset (South region) as the coefficients to construct PRS for the target (North region) [27,50]. Details on selection of sex-specific genes are given in Supplementary Materials (SM) Section SM4. Calculation of the PRS for each of the sex-specific genes was made using the PLINK software. First, we extracted all independent genes associated with longevity of one sex only in the discovery dataset. Second, we removed those genes which are significantly associated with longevity in the other sex in the discovery dataset via PLINK. Each of the genes retained are sex-specific genes; significantly associated with longevity in one sex but not associated with longevity for the other sex. Third, following the method for constructing the standardized PRS developed by Purcell et al. [51], these sets of selected sex-specific genes, which were weighted by their log odds ratios (ORs) from the discovery dataset and summarized into the PRS for each individual in the replication dataset.
To make the PRS more comparable across genders and to improve the interpretability of the results of the logistic regression analyses, we standardized the PRS for each of the identified exclusive groups of genes associated with longevity at different significance levels following the standard approach as expressed in formula (1) below [52].
| (1) |
where is the standardized PRS for individual i with sex s (s = 1 for males and s = 2 for females), is the initial un-standardized PRS for individual i with sex s, and are the mean and the standard deviation of the initial PRS values of all of the male and female individuals of the sample. After rescaling of the z-scores transformation, the standardized PRS have a mean of 0 and a standard deviation of 1.0; this standardization substantially improves comparability across genders [51]. We abbreviate “standardized PRS” as “PRS” in this article to simplify the presentations.
M3. The logistic regression model employed in present study
Note that regression analyses of PRS for males and females separately are not appropriate for adequately quantifying sex differences in genetic associations with longevity, because they use different reference groups for males and females, making the sex-specific estimates not fully compatible. Thus, we estimated logistic regression models including both males and females, in which the binary dependent variable is being a centenarian or middle-age control, and the independent variables are sex, the genetic variant measured by the continuous PRS, and the PRS-sex interaction term, as expressed in Equations (2), (3) below:
| (2) |
where P(x,s)i is the probability of longevity (likelihood of being a centenarian) of the individual i with sex s (s = 1, male; s = 2, female) and genetic propensity for longevity measured by standardized continuous PRS value x; s and x for individual i are denoted as Si and Xi; (Xi*Si) represents the PRS-sex interaction effects; β0 is the intercept; β1, β2, and β3 are the regression coefficients of the independent variables Si and Xi, and (Xi*Si) interaction term, respectively.
We first estimate the , which is the odds(x,s), based on the logistic regression expressed in Equation (2): odds(x,s) = .
We then derive as follows, based on the estimated odds(x,s) as expressed above:
| (3) |
We use 0.01 as the increment to transfer the continuous PRS into discrete numbers of x, which
keep two decimal points. Let Hx denote the highest PRS value included in the regression analysis;
Lx, the lowest PRS value included in the regression analysis;
P(x,s), the probability of longevity among persons of sex s and with the PRS value x, estimated based on the logistic regression model expressed in Equations (2), (3);
AP(s), the average probabilities of longevity among persons of sex s with various PRS values.
| (4) |
The strength of our logistic regression analyses is that the sex-specific estimates are fully compatible, and can be used to quantify sex differences in genetic associations with longevity. Furthermore, given the available datasets, this regression model which includes all individuals of males and females provides substantially more statistical power than performing regression analyses for males and females separately.
Consistent with other published case-control studies of GWAS on longevity based on cross-sectional datasets of centenarians and middle-age controls, we did not include socioeconomic covariates (e.g. education, occupation, etc.) in our logistic regression models, because the socioeconomic factors of the two birth cohorts of centenarians and middle-age controls born 40–60 years apart are not compatible. Instead, in our GWAS regression models, we adjusted for the top two eigenvectors to minimize the effects of population stratification, following the approach adopted in the GWAS literature [53].
Mood [54] demonstrated that, compared with probabilities (P(x,s)), the sex-specific odds(x,s) (=P(x,s)/(1-P(x,s))) and males/females odds ratio (x) (=Odds(x,1)/Odds(x,2)) cannot be interpreted as absolute effects, nor can they be accurately compared across male and female groups, although they are widely used to measure the degree and direction of the associations between two categories within one group. For example, assuming probability of longevity of a good genotype is 0.55 and 0.40 among males and females, respectively, the males/females relative risk ratio is 1.375 (=0.55/0.40), which means that the genotype's effects in males are 37.5 % higher than that in females. However, the sex-specific odds(x,s) calculated using the probability of longevity are 1.222 (=0.55/(1-0.55)) for males and 0.667 (=0.40/(1-0.40)) for females. The males/females odds ratio (x) of females to males is 1.83 (=1.222/0.667), which means that the genotype's effects in males are 83 % higher than that in females, exaggerating the sex differences in the effects of the genotype by 2.22 folds (=83.3 %/37.5 %).
Norton et al. (2018; Page 84) [55] stated (we cite their original statements here): “Although for rare outcomes odds ratios approximate relative risk ratios (which is the ratio of probabilities of the outcome for two groups such as males and females), when the outcomes are not rare, odds ratios always overestimate relative risk ratios, a problem that becomes more acute as the baseline prevalence of the outcome exceeds 10 %. For example, an odds ratio (men/women) of 2.0 could correspond to the situation in which the probability for some event is 1 % for males and 0.5 % for females. An odds ratio (men/women) of 2.0 also could correspond to a probability of an event occurring 50 % for males and 33 % for females, or to a probability of 80 % for males and 67 % for females.” The SM-Table 6 presents the numerical calculations and comparisons between the males/females relative risk ratios and males/females odds ratios of the three examples given by Norton et al. [55] as outlined above and four examples of the PRS values of the 11 male-specific top genes of longevity (P < 10−5 in males, but P > 0.16 in females) estimated using the CLHLS GWAS datasets (see Fig. 1a).
What Norton et al. [55] stated “… when the outcomes are not rare, odds ratios always overestimate relative risk ratios” can be mathematically derived in connection with present study as follows. Denote P(x,s) (s = 1: males, s = 2: females) as probability of longevity associated with PRS value x; males/females relative risk ratio is: P(x,1)/P(x,2); the sex-specific Odds(x,s) is: P(x,s)/(1- P(x,s));
| (5) |
formula (5) indicates that the odds ratio (x) biases the males/females relative risk ratios by a factor of , if the odds ratio is used to quantify the differences between males and females. Thus, the odds ratio cannot be used to accurately quantify the differences in genetic associations between males and females, although they are widely used to measure the degree and direction of the associations between two categories within one group (e.g., within male group or within female group) [[54], [55], [56]]. Therefore, referring to the relevant literature [[54], [55], [56]], we estimated and compared the probabilities of longevity (rather than the odds ratios) of various sex-specific and non-sex-specific genes between males and females, in order to accurately quantify the sex differences in genetic associations with longevity.
To address the concern about whether our analyses using logistic regression may bias the estimates of probability of longevity due to the larger sample size of female centenarians compared to their male counterparts, which is common in all studies including centenarians, we conducted additional analysis based on randomly selected female centenarian/control samples with exactly the same size as the male centenarian/control samples. As presented and discussed in details insection 3.5, our additional analyses, with exactly the same sample sizes of the male and female centenarians and controls demonstrated similar results as was found from the analyses using the actual total female and male samples. Thus, our analyses using logistic regression did not bias the estimates of probability of longevity.
M4. Bio-demographic analyses on female to male ratios of relative benefits due to sex-specific genes of longevity
Referring to Fig. 1a and b for an intuitive understanding, let A(s) denote the sex-specific relative benefits (compared with the other-sex) among individuals of sex s (s = 1: males; s = 2: females), due to gaining the positive joint effects of higher PRS (x > C) of the sex-specific genes of longevity; B(s) denotes the sex-specific relative benefits among individuals of sex s, due to avoiding the negative joint effects of lower PRS (x < C) of the other-sex-specific genes of longevity; where C is the PRS value at which the probabilities of longevity in males and females are equal to each other (i.e., the male and female curves cross at x = C). The formulas for estimating the A(s) and B(s) are presented and discussed below.
Female to male ratio of relative benefits due to a group of male-specific genes of longevity.
Referring to Fig. 1a, the triangle shaped area A(1) above the female line and below the male line illustrates the male relative benefits in probability of longevity due to gaining the positive joint effects of higher PRS (x > C) of a group of male-specific genes of longevity; A(1) is the sum of differences in probabilities of longevity between males and females, all with higher PRS (x > C) of the male-specific genes of longevity:
| (6) |
On the other hand, triangle area B(2) in Fig. 1a denotes female relative benefits in probability of longevity due to avoiding the negative joint effects of lower PRS (x < C) of a group of male-specific genes of longevity; B(2) is the sum of differences in probabilities of longevity between females and males, all with lower PRS (x < C) of the male-specific genes of longevity:
| (7) |
| (8) |
FM1 is the female to male ratio of relative benefits due to a group of male-specific genes of longevity, namely, the ratio of females' relative benefits due to avoiding the negative joint effects of lower PRS (x < C) of the male-specific genes of longevity (B(2)) to the males’ relative benefits due to gaining the positive joint effects of higher PRS (x > C) of the same group of male-specific genes of longevity (A(1)). If FM1 is larger (or smaller) than one, it indicates that the relative benefits in probability of longevity due to a group of male-specific genes of longevity are higher (or lower) in females than in males.
Female to male ratio of relative benefits due to a group of female-specific genes of longevity.
Referring to Fig. 1b, the triangle area A(2) denotes female relative benefits in probability of longevity, due to gaining the positive joint effects of higher PRS (x > C) of female-specific genes of longevity; A(2) is the sum of differences in probabilities of longevity between females and males, all with higher PRS (x > C) of the female-specific genes of longevity:
| (9) |
Referring to Fig. 1b, the triangle area B(1) denotes male relative benefits in probability of longevity due to avoiding the negative joint effects of lower PRS (x < C) of the female-specific genes of longevity, B(1) is the sum of differences in probabilities of longevity between males and females, all with lower PRS (x < C) of the female-specific genes of longevity:
| (10) |
| (11) |
FM2 is the female to male ratio of relative benefits due to a group of female-specific genes of longevity, namely, the ratio of females' relative benefits due to gaining the positive joint effects of higher PRS (x > C) of the female-specific genes of longevity (A(2)) to the males’ relative benefits due to avoiding the negative joint effects of lower PRS (x < C) of the same female-specific genes of longevity (B(1)). If FM2 is larger (or smaller) than one, it indicates that the relative benefits due to the female-specific genes of longevity are higher (or lower) in females than in males.
M5. The genome-wide Complex Trait Analysis (GCTA) method used in present study
Chip-based heritability was estimated for each sex based on the CLHLS GWAS and healthy aging candidate genes datasets respectively in present study, using the GCTA restricted maximum likelihood (REML) method as implemented in GCTA version 1.92.0beta3 [30]. SNP variants were linkage disequilibrium pruned using PLINK2 and the flag --indep-pairwise 50 10 0.1. The genetic relationship matrix (GRM) was constructed and the heritability was calculated with flags, using the GCTA. In order to transform the estimated heritability from the observed scale to liability scale, we specified the flag of prevalence 0.0002 for males and prevalence 0.0006 for females, according to the life expectations at birth, as reported in “World Population Prospects 2019” released by United Nations Population Division.
Data availability statement
The data reported in this paper have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences [57,58] (https://ngdc.cncb.ac.cn/omix:accessionnoOMIX003054). All data requests should be submitted to the corresponding author for consideration.
Data sources: The sex-specific genes outlined in (1), (2), (3), (4) were identified using CLHLS GWAS datasets, and their more detailed information are referred to Table 1, Table 2 and eTables 4-5 of the reference23. The sex-specific genes outlined in (5), (6) are identified in present study using the CLHLS GWAS dataset and their more detailed information are presented in SM-Table 3a and SM-Table 3b. The sex-specific genes outlined in (7), (8) are identified in present study using the CLHLS healthy aging candidate genes datasets and their more detailed information are presented in SM-Table 4a and SM-Table 4b. Notes: (a) Each of the eight groups of sex-specific genes of longevity listed in this Table reaches a jointly-significant level of P < 10−8 in one sex but not-jointly-significant in the other sex (P > 0.05) 23 (ref. SM-Table 5 for details); (b) The estimates presented in this table are based on the logistic regressions including continuous standardized PRS, sex and (PRS x sex) interaction term (ref. Equations (2), (3) in the Methods section M3); (c) The sex-specific average probabilities of longevity among persons of sex s with various PRS values presented in this table are estimated using formula (4) in Methods section M3; (d) The P values of the sex differences presented in the last column of this table are estimated using the bootstrap method (the number of bootstrap replications is set to 2000) of the statistical software STATA 12.0.
CRediT authorship contribution statement
Yi Zeng: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Visualization, Writing - original draft, Writing - review & editing, Resources, Supervision, Validation. Huashuai Chen: Writing - review & editing. Xiaomin Liu: Visualization, Writing - review & editing. Zijun Song: Investigation, Writing - review & editing. Yao Yao: Investigation. Xiaoyan Lei: Funding acquisition, Investigation. Xiaozhen Lv: Investigation. Lingguo Cheng: Investigation. Zhihua Chen: Investigation. Chen Bai: Investigation. Zhaoxue Yin: Investigation. Yuebin Lv: Investigation. Jiehua Lu: Investigation. Jianxin Li: Investigation. Kenneth C. Land: Investigation. Anatoliy Yashin: Investigation. Angela M. O'Rand: Investigation. Liang Sun: Investigation. Ze Yang: Investigation, Writing - review & editing. Wei Tao: Investigation, Writing - review & editing. Jun Gu: Investigation. William Gottschalk: Investigation. Qihua Tan: Investigation. Kaare Christensen: Investigation. Therese Hesketh: Investigation. Xiao-Li Tian: Investigation. Huanming Yang: Investigation. Viviana Egidi: Investigation. Graziella Caselli: Investigation. Jean-Marie Robine: Investigation. Huali Wang: Investigation. Xiaoming Shi: Investigation, Supervision. James W. Vaupel: Investigation. Michael W. Lutz: Investigation. Chao Nie: Data curation, Investigation, Supervision, Visualization, Writing - review & editing. Junxia Min: Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Yuzhi Liu, Chunyuan Zhang, Yun Zhou and Zhenzhen Zheng from Peking University; Zhenyu Xiao, Liqun Tao, Qin Xu and Ye Yuan from Chinese Center for Aging Science Research; and Jie Zhan from China Social Science Academy for their contributions to the Chinese Longitudinal Healthy Longevity Study (CLHLS) data collections, and we thank all interviewees and their families for their voluntary participation in the CLHLS study. We appreciate very much for Dr. Xuxi Zhang's help and thoughtful comments in the revision & resubmission of this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23691.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Van Oyen H., Nusselder W., Jagger C., Kolip P., Cambois E., Robine J.M. Gender differences in healthy life years within the EU: an exploration of the “health–survival” paradox. Int. J. Publ. Health. 2013;58:143–155. doi: 10.1007/s00038-012-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dicker D., Nguyen G., Abate D., Abate K.H., Abay S.M., Abbafati C., Global .. S.A. Belay. regional, and national age-sex-specific mortality and life expectancy, 1950–2017: a systematic analysis for the Global Burden of Disease Study 2017. The lancet. 2018;392:1684–1735. doi: 10.1016/S0140-6736(18)31891-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrán-Sánchez H., Finch C.E., Crimmins E.M. Twentieth century surge of excess adult male mortality. Proc. Natl. Acad. Sci. USA. 2015;112:8993–8998. doi: 10.1073/pnas.1421942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarulli V., Jones J.A., Oksuzyan A., Lindahl-Jacobsen R., Christensen K., Vaupel J.W. Women live longer than men even during severe famines and epidemics. Proc. Natl. Acad. Sci. USA. 2018;115:E832–E840. doi: 10.1073/pnas.1701535115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macintyre K. In: Famine Demography: Perspectives from the Past and Present. Allen T., et al., editors. Oxford Univ Press; Oxford: 2002. Famine and the female mortality advantage; pp. 240–260. [Google Scholar]
- 6.Watkins S.C., Menken J. Famines in historical perspective. Popul. Dev. Rev. 1985:647–675. [Google Scholar]
- 7.Robine J.M., Michel J.P., Herrmann F.R. Excess male mortality and age-specific mortality trajectories under different mortality conditions: a lesson from the heat wave of summer 2003. Mechanisms of ageing and development. 2012;133:378–386. doi: 10.1016/j.mad.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 8.China CDC Weekly Reports. 2020. http://weekly.chinacdc.cn/index.htm [Google Scholar]
- 9.KCDC (Korean Centers for Disease Control Prevention) 2020. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030
- 10.D'Arminio Monforte A., Tavelli A., Bai F., Tomasoni D., Falcinella C., Castoldi R., Marchetti .G. Declining mortality rate of hospitalised patients in the second wave of the COVID-19 epidemics in Italy: risk factors and the age-specific patterns. Life. 2021;11:979. doi: 10.3390/life11090979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudel C., Riffe T., Acosta E., van Raalte A., Strozza C., Myrskylä M. Monitoring trends and differences in COVID-19 case-fatality rates using decomposition methods: contributions of age structure and age-specific fatality. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudharsanan N., Didzun O., Bärnighausen T., Geldsetzer P. The contribution of the age distribution of cases to COVID-19 case fatality across countries: a nine-country demographic study. Annals of internal medicine. 2020;173:714–720. doi: 10.7326/M20-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Y., Feng Q., Hesketh T., Christensen K., Vaupel J.W. Survival, disabilities in activities of daily living, and physical and cognitive functioning among the oldest-old in China: a cohort study. Lancet. 2017;389:1619–1629. doi: 10.1016/S0140-6736(17)30548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alesina A., Giuliano P., Nunn N. Traditional agricultural practices and the sex ratio today. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adjei N.K., Brand T., Zeeb H. Gender inequality in self-reported health among the elderly in contemporary welfare countries: a cross-country analysis of time use activities, socioeconomic positions and family characteristics. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deelen J., Beekman M., Uh H.W., Broer L., Ayers K.L., Tan Q., Slagboom .P.E. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum. Mol. Genet. 2014;23:4420–4432. doi: 10.1093/hmg/ddu139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman A.B., Walter S., Lunetta K.L., Garcia M.E., Slagboom P.E., Christensen K., Murabito .J.M. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. 2010;65:478–487. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebastiani P., Solovieff N., DeWan A.T., Walsh K.M., Puca A., Hartley S.W., Perls .T.T. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y., Nie C., Min J., Liu X., Li M., Chen H., Vaupel .J.W. Novel loci and pathways significantly associated with longevity. Sci. Rep. 2016;6 doi: 10.1038/srep21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y., Nie C., Min J., Chen H., Liu X., Ye R., Vaupel .J. Sex differences in genetic associations with longevity. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilks W.P., Abbott J.K., Morrow E.H. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet. 2014;30:453–463. doi: 10.1016/j.tig.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Nebel A., Flachsbart F., Till A., Caliebe A., Blanché H., Arlt A., Schreiber .S. A functional EXO1 promoter variant is associated with prolonged life expectancy in centenarians. Mechanisms of ageing and development. 2009;130:691–699. doi: 10.1016/j.mad.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Willcox B.J., Donlon T.A., He Q., Chen R., Grove J.S., Yano K., Curb .J.D. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anselmi C.V., Malovini A., Roncarati R., Novelli V., Villa F., Condorelli G., Puca .A.A. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Annu. Rev. Biomed. Eng. 2009;25:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Wang W.J., Cao H., Lu J., Wu C., Hu F.Y., Tian X.L. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum. Mol. Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji J.S., Liu L., Shu C., Yan L.L., Zeng Y. Sex difference and interaction of SIRT1 and FOXO3 candidate longevity genes on life expectancy: a 10-year prospective longitudinal cohort study. J. Gerontol.: Series A. 2022;77:1557–1563. doi: 10.1093/gerona/glab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Euesden J., Lewis C.M., Prsice P.F. O'Reilly. Polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K., Taskesen E., Van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snow A.D., Cummings J.A., Lake T. The unifying hypothesis of Alzheimer's disease: heparan sulfate proteoglycans/glycosaminoglycans are key as first hypothesized over 30 years ago. Front. Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.710683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera S., Garvert L., Fuellen G., Nauck M., Völzke H., Völker U., Grabe H.J. The genetic predisposition to longevity acts through behavioral phenotypes in females. Eur. Neuropsychopharmacol. 2021;45:1–14. doi: 10.1016/j.euroneuro.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Wachter K.W., Evans S.N., Steinsaltz D. The age-specific force of natural selection and biodemographic walls of death. Proc. Natl. Acad. Sci. USA. 2013;110:10141–10146. doi: 10.1073/pnas.1306656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim P.S., McQueen J.S., Coxworth J.E., Hawkes K. Grandmothering drives the evolution of longevity in a probabilistic model. J. Theor. Biol. 2014;353:84–94. doi: 10.1016/j.jtbi.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Perls T.T., Alpert L., Fretts R.C. Middle-aged mothers live longer. Nature. 1997;389 doi: 10.1038/38148. [DOI] [PubMed] [Google Scholar]
- 35.Smith K.R., Mineau G.P., Bean L.L. Fertility and post‐reproductive longevity. Soc. Biol. 2002;49:185–205. [PubMed] [Google Scholar]
- 36.Yi Z., Vaupelly J.W. Association of late childbearing with healthy longevity among the oldest-old in China. Popul. Stud. 2004;58:37–53. doi: 10.1080/0032472032000175437. [DOI] [PubMed] [Google Scholar]
- 37.Perls T.T., Fretts R.C. Why women live longer than men-what gives women the extra years? Sci. Am. 1998;2:100–103. [Google Scholar]
- 38.Della Torre S., Mitro N., Meda C., Lolli F., Pedretti S., Barcella M., Maggi .A. Short-term fasting reveals amino acid metabolism as a major sex-discriminating factor in the liver. Cell Metabol. 2018;28:256–267. doi: 10.1016/j.cmet.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 40.Goetzl E.J., Huang M.C., Kon J., Patel K., Schwartz J.B., Fast K., Longo .D.L. Gender specificity of altered human immune cytokine profiles in aging. Faseb. J. 2010;24:3580. doi: 10.1096/fj.10-160911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hewagama A., Patel D., Yarlagadda S., Strickland F.M., Richardson B.C. Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Gene Immun. 2009;10:509–516. doi: 10.1038/gene.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds-Peterson C., Xu J., Zhao N., Cruse C., Yonel B., Trasorras C., Selleck S. Heparan sulfate structure affects autophagy, lifespan, responses to oxidative stress, and cell degeneration in Drosophila parkin mutants. G3: Genes, Genomes, Genetics. 2020;10:129–141. doi: 10.1534/g3.119.400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tower J., Arbeitman M. The genetics of gender and life span. Journal of biology. 2009;8:1–3. doi: 10.1186/jbiol141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis E.J., Lobach I., Dubal D.B. Female XX sex chromosomes increase survival and extend lifespan in aging mice. Aging Cell. 2019;18 doi: 10.1111/acel.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumitrescu L., Barnes L.L., Thambisetty M., Beecham G., Kunkle B., Bush W.S., Hohman .T.J. Sex differences in the genetic predictors of Alzheimer's pathology. Brain. 2019;142:2581–2589. doi: 10.1093/brain/awz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prokopenko D., Hecker J., Kirchner R., Chapman B.A., Hoffman O., Mullin K., Tanzi .R.E. Identification of novel Alzheimer's disease loci using sex-specific family-based association analysis of whole-genome sequence data. Sci. Rep. 2020;10:5029. doi: 10.1038/s41598-020-61883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jameson J.L., Longo D.L. Precision medicine—personalized, problematic, and promising. Obstetrical gynecological survey. 2015;70:612–614. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 48.Coale A.J., Li S. The effect of age misreporting in China on the calculation of mortality rates at very high ages. Demography. 1991;28:293–301. [PubMed] [Google Scholar]
- 49.Wang Z., Zeng Y., Jeune B., Vaupel J.W. Age validation of Han Chinese centenarians. Genus. 1998:123–141. [PubMed] [Google Scholar]
- 50.Xu S., Yin X., Li S., Jin W., Lou H., Yang L., Jin .L. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am. J. Hum. Genet. 2009;85:762–774. doi: 10.1016/j.ajhg.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis C.M., Vassos E. Prospects for using risk scores in polygenic medicine. Genome Med. 2017;9:1–3. doi: 10.1186/s13073-017-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price A.L., Butler J., Patterson N., Capelli C., Pascali V.L., Scarnicci F., Hirschhorn .J.N. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mood C. Logistic regression: why we cannot do what we think we can do, and what we can do about it. Eur. Socio Rev. 2010;26:67–82. [Google Scholar]
- 55.Norton E.C., Dowd B.E., Maciejewski M.L. Odds ratios—current best practice and use. JAMA. 2018;320:84–85. doi: 10.1001/jama.2018.6971. [DOI] [PubMed] [Google Scholar]
- 56.Hoetker G. The use of logit and probit models in strategic management research: Critical issues. Strat. Manag. J. 2007;28:331–343. [Google Scholar]
- 57.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Zhao .W. The genome sequence archive family: toward explosive data growth and diverse data types. Dev. Reprod. Biol. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 2022;50:D27–D38. doi: 10.1093/nar/gkab951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences [57,58] (https://ngdc.cncb.ac.cn/omix:accessionnoOMIX003054). All data requests should be submitted to the corresponding author for consideration.
Data sources: The sex-specific genes outlined in (1), (2), (3), (4) were identified using CLHLS GWAS datasets, and their more detailed information are referred to Table 1, Table 2 and eTables 4-5 of the reference23. The sex-specific genes outlined in (5), (6) are identified in present study using the CLHLS GWAS dataset and their more detailed information are presented in SM-Table 3a and SM-Table 3b. The sex-specific genes outlined in (7), (8) are identified in present study using the CLHLS healthy aging candidate genes datasets and their more detailed information are presented in SM-Table 4a and SM-Table 4b. Notes: (a) Each of the eight groups of sex-specific genes of longevity listed in this Table reaches a jointly-significant level of P < 10−8 in one sex but not-jointly-significant in the other sex (P > 0.05) 23 (ref. SM-Table 5 for details); (b) The estimates presented in this table are based on the logistic regressions including continuous standardized PRS, sex and (PRS x sex) interaction term (ref. Equations (2), (3) in the Methods section M3); (c) The sex-specific average probabilities of longevity among persons of sex s with various PRS values presented in this table are estimated using formula (4) in Methods section M3; (d) The P values of the sex differences presented in the last column of this table are estimated using the bootstrap method (the number of bootstrap replications is set to 2000) of the statistical software STATA 12.0.