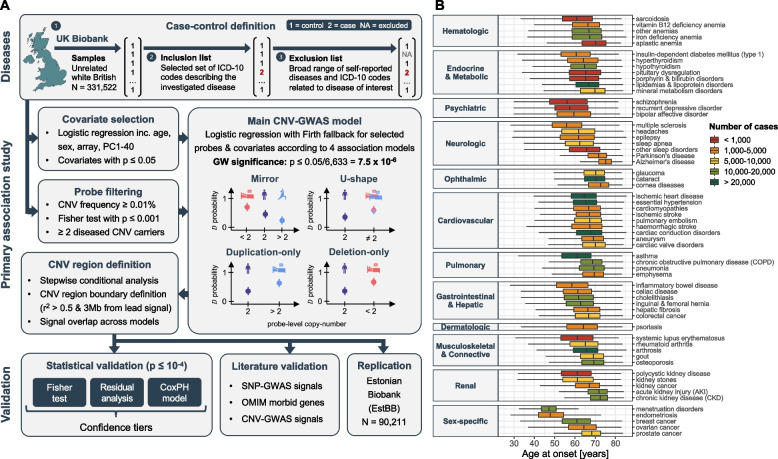
Fig. 1.
Overview of the study. A Schematic representation of the analysis workflow. Diseases: For each of the 60 investigated diseases, 331,552 unrelated white British individuals were divided into three subsets: controls (encoded as 1; step 1), cases with the disease (encoded as 2; step 2), and a subset of individuals who were excluded because they had conditions similar but not identical to the disease (encoded as NA; step 3). Primary association study: Disease-specific relevant covariates were selected. Probes were pre-filtered based on copy-number variant (CNV) frequency, Fisher test association p-value, and presence of ≥ 2 diseased carriers. Disease- and model-specific covariates and probes were used to generate tailored genome-wide CNV association scans (CNV-GWASs) based on Firth fallback logistic regression according to a mirror, U-shape, duplication-only (i.e., considering only duplications), and deletion-only (i.e., considering only deletions) models. Independent lead signals were identified through stepwise conditional analysis and CNV regions were defined based on probe correlation and merged across models. Validation: Statistical validation methods (i.e., Fisher test, residuals regression, and Cox proportional hazards model (CoxPH)) were used to rank associations in confidence tiers. Literature validation approaches leverage data from independent studies to corroborate that genetic perturbation (single-nucleotide polymorphisms (SNP), rare variants from the OMIM database, or CNVs) in the region are linked to the disease. Independent replication in the Estonian Biobank. B Age of onset for the 60 assessed diseases, grouped based on ICD-10 chapters and colored according to case count. Data are represented as boxplots; outliers are not shown