Abstract
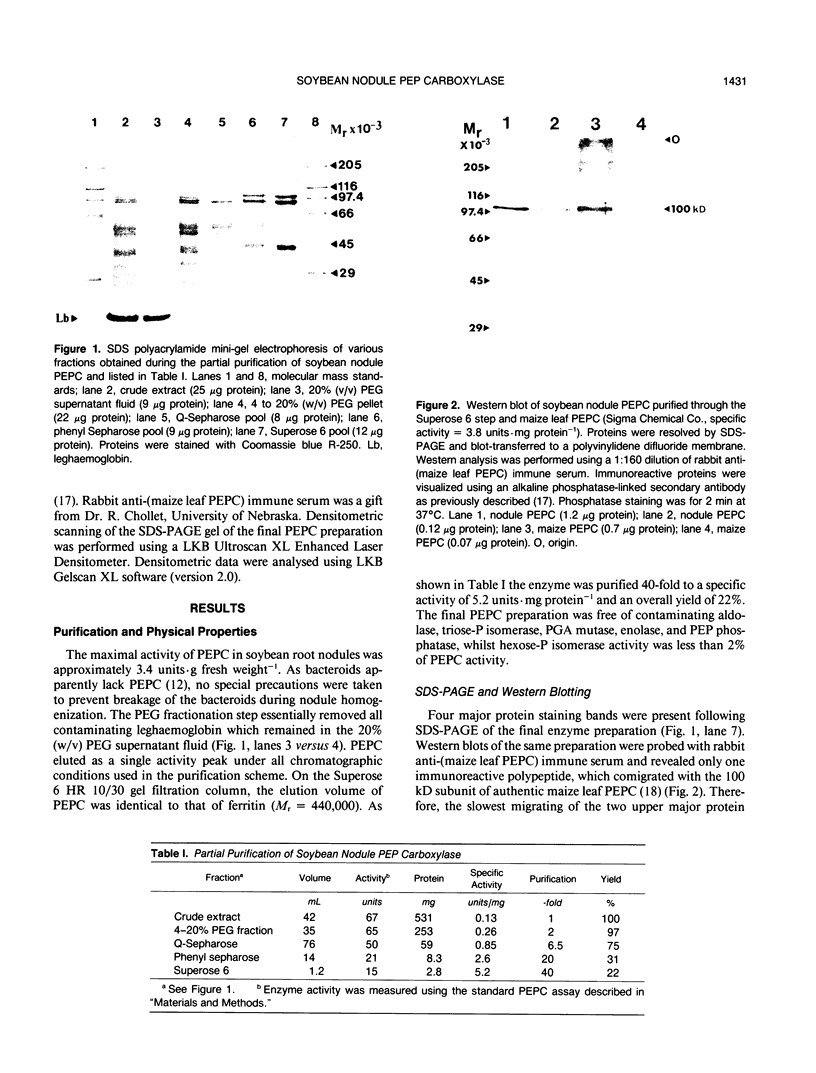
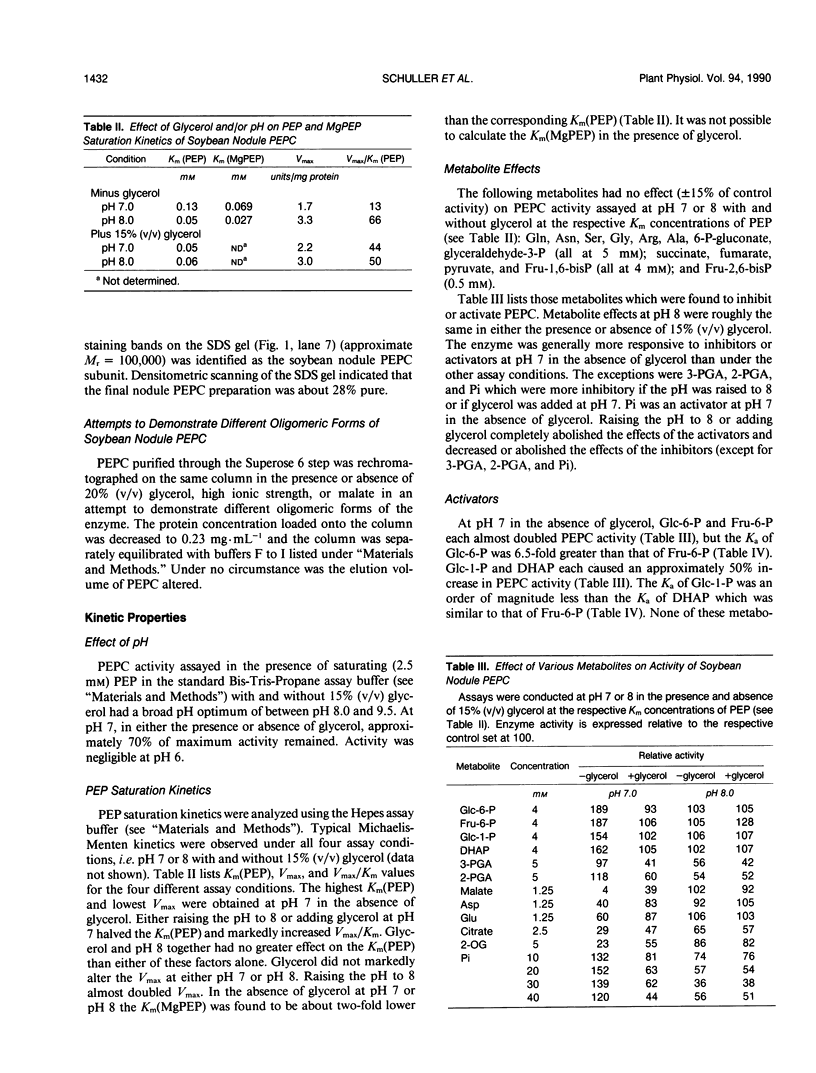
Phosphoenolpyruvate carboxylase (PEPC) was purified 40-fold from soybean (Glycine max L. Merr.) nodules to a specific activity of 5.2 units per milligram per protein and an estimated purity of 28%. Native and subunit molecular masses were determined to be 440 and 100 kilodaltons, respectively, indicating that the enzyme is a homotetramer. The response of enzyme activity to phosphoenolpyruvate (PEP) concentration and to various effectors was influenced by assay pH and glycerol addition to the assay. At pH 7 in the absence of glycerol, the Km (PEP) was about twofold greater than at pH 7 in the presence of glycerol or at pH 8. At pH 7 or pH 8 the Km (MgPEP) was found to be significantly lower than the respective Km (PEP) values. Glucose-6-phosphate, fructose-6-phosphate, glucose-1-phosphate, and dihydroxyacetone phosphate activated PEPC at pH 7 in the absence of glycerol, but had no effect under the other assay conditions. Malate, aspartate, glutamate, citrate, and 2-oxoglutarate were potent inhibitors of PEPC at pH 7 in the absence of glycerol, but their effectiveness was decreased by raising the pH to 8 and/or by adding glycerol. In contrast, 3-phosphoglycerate and 2-phosphoglycerate were less effective inhibitors at pH 7 in the absence of glycerol than under the other assay conditions. Inorganic phosphate (up to 20 millimolar) was an activator at pH 7 in the absence of glycerol but an inhibitor under the other assay conditions. The possible significance of metabolite regulation of PEPC is discussed in relation to the proposed functions of this enzyme in legume nodule metabolism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Canellas P. F., Wedding R. T. Substrate and metal ion interactions in the NAD+ malic enzyme from cauliflower. Arch Biochem Biophys. 1980 Jan;199(1):259–264. doi: 10.1016/0003-9861(80)90279-9. [DOI] [PubMed] [Google Scholar]
- Christeller J. T., Laing W. A., Sutton W. D. Carbon Dioxide Fixation by Lupin Root Nodules: I. Characterization, Association with Phosphoenolpyruvate Carboxylase, and Correlation with Nitrogen Fixation during Nodule Development. Plant Physiol. 1977 Jul;60(1):47–50. doi: 10.1104/pp.60.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff S. M., Lefebvre D. D., Plaxton W. C. Purification and Characterization of a Phosphoenolpyruvate Phosphatase from Brassica nigra Suspension Cells. Plant Physiol. 1989 Jun;90(2):734–741. doi: 10.1104/pp.90.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel D. W., Jackson W. A. Ion balance, uptake, and transport processes in n(2)-fixing and nitrate- and urea-dependent soybean plants. Plant Physiol. 1982 Jan;69(1):171–178. doi: 10.1104/pp.69.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job D., Cochet C., Dhien A., Chambaz E. M. A rapid method for screening inhibitor effects: determination of I50 and its standard deviation. Anal Biochem. 1978 Jan;84(1):68–77. doi: 10.1016/0003-2697(78)90484-0. [DOI] [PubMed] [Google Scholar]
- King B. J., Layzell D. B., Canvin D. T. The role of dark carbon dioxide fixation in root nodules of soybean. Plant Physiol. 1986 May;81(1):200–205. doi: 10.1104/pp.81.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton G. A., Fewson C. A., Wilkins M. B., Nimmo H. G. Purification, oligomerization state and malate sensitivity of maize leaf phosphoenolpyruvate carboxylase. Biochem J. 1989 Jul 15;261(2):349–355. doi: 10.1042/bj2610349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. S., Boylan K. L., Vance C. P. Alfalfa Root Nodule Carbon Dioxide Fixation : III. Immunological Studies of Nodule Phosphoenolpyruvate Carboxylase. Plant Physiol. 1987 Jun;84(2):501–508. doi: 10.1104/pp.84.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G. B., Plaxton W. C. Purification and characterization of cytosolic aldolase from carrot storage root. Biochem J. 1990 Jul 1;269(1):133–139. doi: 10.1042/bj2690133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. B., Evans H. J. Phosphoenolpyruvate carboxylase from soybean nodule cytosol. Evidence for isoenzymes and kinetics of the most active component. Biochim Biophys Acta. 1979 Apr 12;567(2):445–452. doi: 10.1016/0005-2744(79)90130-x. [DOI] [PubMed] [Google Scholar]
- Plaxton W. C. Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from castor-oil-plant endosperm and leaf. Eur J Biochem. 1989 May 1;181(2):443–451. doi: 10.1111/j.1432-1033.1989.tb14745.x. [DOI] [PubMed] [Google Scholar]
- Podestá F. E., Andreo C. S. Maize leaf phosphoenolpyruvate carboxylase : oligomeric state and activity in the presence of glycerol. Plant Physiol. 1989 Jun;90(2):427–433. doi: 10.1104/pp.90.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibach P. H., Streeter J. G. Metabolism of C-labeled photosynthate and distribution of enzymes of glucose metabolism in soybean nodules. Plant Physiol. 1983 Jul;72(3):634–640. doi: 10.1104/pp.72.3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl L., Vance C. P., Pedersen W. B. Products of Dark CO(2) Fixation in Pea Root Nodules Support Bacteroid Metabolism. Plant Physiol. 1990 May;93(1):12–19. doi: 10.1104/pp.93.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller K. A., Plaxton W. C., Turpin D. H. Regulation of Phosphoenolpyruvate Carboxylase from the Green Alga Selenastrum minutum: Properties Associated with Replenishment of Tricarboxylic Acid Cycle Intermediates during Ammonium Assimilation. Plant Physiol. 1990 Aug;93(4):1303–1311. doi: 10.1104/pp.93.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter J. G. Carbohydrate, organic Acid, and amino Acid composition of bacteroids and cytosol from soybean nodules. Plant Physiol. 1987 Nov;85(3):768–773. doi: 10.1104/pp.85.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin D. H., Botha F. C., Smith R. G., Feil R., Horsey A. K., Vanlerberghe G. C. Regulation of Carbon Partitioning to Respiration during Dark Ammonium Assimilation by the Green Alga Selenastrum minutum. Plant Physiol. 1990 May;93(1):166–175. doi: 10.1104/pp.93.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C. P., Stade S. Alfalfa Root Nodule Carbon Dioxide Fixation : II. Partial Purification and Characterization of Root Nodule Phosphoenolpyruvate Carboxylase. Plant Physiol. 1984 May;75(1):261–264. doi: 10.1104/pp.75.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G. C., Schuller K. A., Smith R. G., Feil R., Plaxton W. C., Turpin D. H. Relationship between NH(4) Assimilation Rate and in Vivo Phosphoenolpyruvate Carboxylase Activity : Regulation of Anaplerotic Carbon Flow in the Green Alga Selenastrum minutum. Plant Physiol. 1990 Sep;94(1):284–290. doi: 10.1104/pp.94.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedding R. T., Black M. K., Meyer C. R. Inhibition of phosphoenolpyruvate carboxylase by malate. Plant Physiol. 1990 Feb;92(2):456–461. doi: 10.1104/pp.92.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedding R. T., Rustin P., Meyer C. R., Black M. K. Kinetic studies of the form of substrate bound by phosphoenolpyruvate carboxylase. Plant Physiol. 1988 Dec;88(4):976–979. doi: 10.1104/pp.88.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]