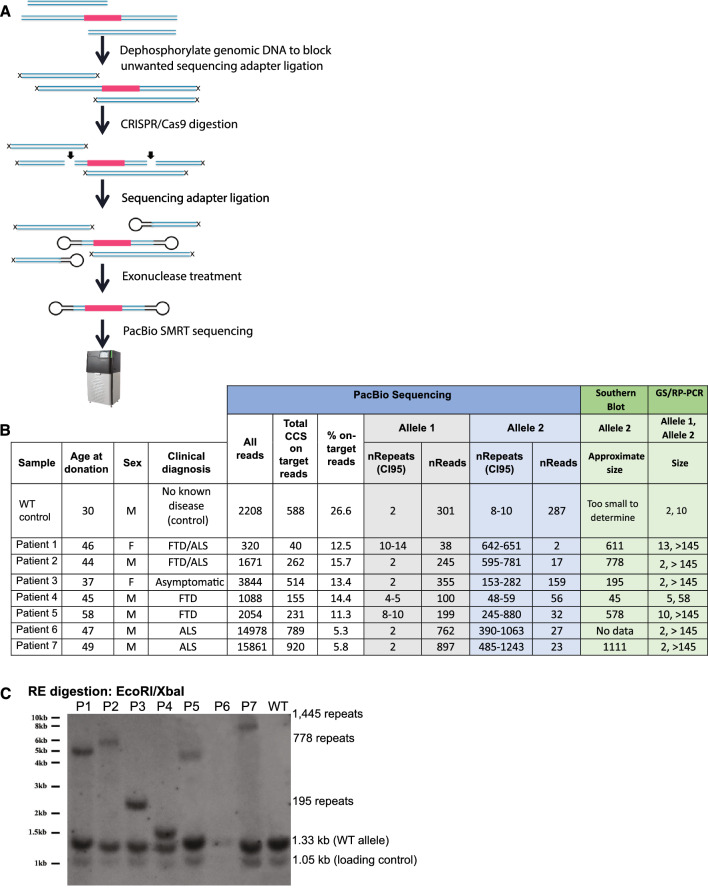
Figure 1.
Pacific Biosciences (PacBio) single-molecule sequencing to determine the C9orf72 repeat size in 8 iPSC lines. (A) Schematic of the pipeline used to generate the library for single-molecule sequencing. We excise the repeat region (red) from high-molecular-weight DNA using CRISPR and guide RNAs flanking the repeat regions (arrows). We then seal the CRISPR-generated double-strand breaks by ligating in sequencing adapters. Subsequent exonuclease treatment results in an enrichment for the excised repeat region, which is sealed at both ends. The enriched repeat regions are then subjected to PacBio SMRT sequencing. Because the sequenced molecules are circular, the sequencing reaction can read through them more than once, which increases the accuracy of the sequencing data. Barcoding allows us to multiplex samples to reduce sequencing costs. (B) We sequenced 3–5 µg of DNA from 1 WT-control iPSC line and 7 iPSC lines from patients harboring expansions of the C9orf72 repeat. Allele-specific SNPs allowed us to distinguish the repeat regions from the two C9orf72 alleles (Allele 1 and Allele 2) in each cell line. On-target reads are reads that sequenced the entire excised region (including the repeat region and flanking DNA) 3 times or more (> 3 pass criteria). Within these reads, we counted the number of GGCCCC repeats starting right after an anchor (CGCCC) 5′ to the repeat region. Repeat lengths and associated read counts are reported for each allele of each cell line and compared to repeat length estimated by Southern blot and GS/RP-PCR. Repeat lengths estimated by Southern blot were comparable to mean repeat lengths determined by single-molecule PacBio sequencing, while GS/RP-PCR could not determine repeat lengths > 145. (C) Southern blot of nuclear DNA from WT-control and patient iPSCs listed in (B). After EcoR/XbaI digestion, a loading control fragment (1.05 kb), WT fragment (1.33 kb) and fragments with repeat expansions of various lengths were detected. Southern blot required 20 µg of input DNA (vs. 3–5 µg input for PacBio sequencing) and a sample with 14 µg (P6) failed detection, demonstrating the insensitivity of Southern blot.