Figure 3.
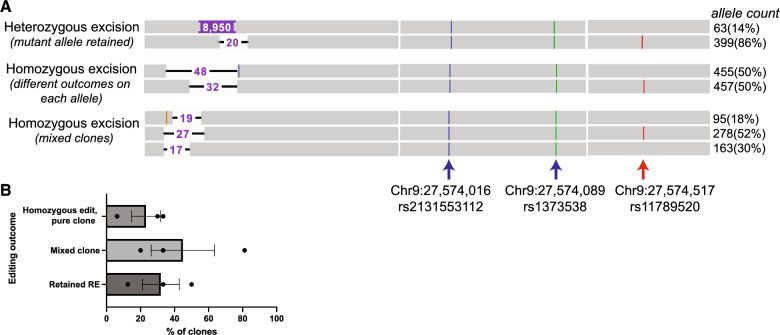
Single molecule sequencing can detect mixed and unedited iPSC clones. (A) Examples of editing outcomes viewed in Integrative Genomics Viewer after sorting and cloning single cells from a pool of edited cells. Each horizontal grey bar represents a single sequenced molecule. Alleles can be distinguished by the phased (red arrow) SNP on the WT allele, but not by homozygous SNPs (blue and green arrow) that differ from the reference genome but are shared by both alleles. SNPs are identified by their unique chromosomal position in GRCh38 and reference SNP cluster number (rsID). We attempted to remove the repeat expansion with CRISPR editing. One clone shows a heterozygous excision with a deletion of 20 nucleotides (NTs) on the WT allele and retention of the C9orf72 repeat expansion (repeats can be variable; 8950 nucleotides or 1491 repeats shown) on the mutant allele. Another clone harbors a homozygous excision with equal read counts across two editing outcomes (48 NT excision on the mutant allele, 32 NT excision on the WT allele). An impure clone shows multiple editing outcomes: a 27 NT excision of the WT allele (52% of sequencing reads), and a 19 NT or 17 NT excision from the mutant allele. The 17 NT excision is twice as abundant as the 19 NT excision in this pool, indicating it is the dominant clone. (B) Dual gRNA excision of the repeat expansion across two cell lines with ~ 200 and ~ 1400 repeats in three independent experiments show that editing outcome detected by single-molecule sequencing of clones detects retained repeat expansions (RE) and mixed clones at a high frequency. Error bar = SEM.