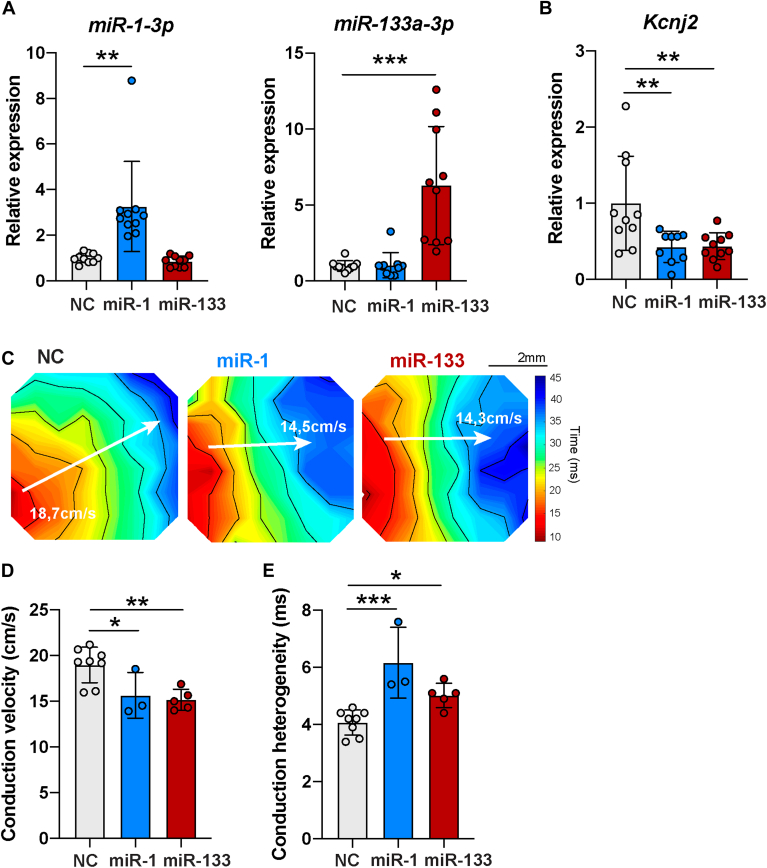
Figure 4.
miR-1-3p and miR-133a-3p transfection in neonatal rat ventricular myocytes (NRVMs) leads to arrhythmogenic features. A: Relative expression of miR-1-3p and miR-133a-3p in NRVMs transfected with negative control (NC), miR-1-3p and miR-133a-3p, measured by real-time quantitative polymerase chain reaction. Data are mean ± SD. Negative control (NC): n = 10; miR-1-3p: n = 10; miR-133a-3p: n = 10 from 3 independent NRVM isolations. Ordinary 1-way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test for miR-1-3p. Kruskal-Wallis followed by Dunnett's multiple comparisons test for miR-133a-3p. Primer sequences are included in Supplemental Tables 3 and 4. B: Relative expression of Kcnj2, gene encoding for Kir2.1, in NRVMs transfected with NC, miR-1-3p, and miR-133a-3p groups, measured by real-time quantitative polymerase chain reaction. Data are mean ± SD. NC: n = 10; miR-1-3p: n = 9; miR-133a-3p: n = 10 from 3 independent NRVM isolation. Ordinary 1-way ANOVA followed by Dunnett's multiple comparisons test. Outlier test (ROUT Q = 1%) identified 1 outlier point in miR-1-3p group that was removed. Primer sequences are included in Supplemental Tables 3 and 4. C: Representative activation maps obtained after electrical mapping of NRVMs transfected with NC, miR-1-3p, or miR-133a-3p. Colors indicate activation times according to the scale at right. Isochrones, 5 ms. White arrows indicate where conduction velocity (CV) was measured. CV is indicated for each activation map in white. D, E: CV and conduction heterogeneity in paced cardiac monolayers after transfection with NC, miR-1-3p, or miR-133a-3p. Data are mean ± SD, n ≥ 3 monolayers from 1 NRVM isolation. Ordinary 1-way ANOVA followed by Dunnett's multiple comparisons test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.