SUMMARY
Zika virus (ZIKV) is an emerging pathogen that causes devastating congenital defects. The overlapping epidemiology and immunologic cross-reactivity between ZIKV and dengue virus (DENV) pose complex challenges to vaccine design, given the potential for antibody-dependent enhancement of disease. Therefore, classification of ZIKV-specific antibody targets is of notable value. From a ZIKV-infected rhesus macaque, we identify ZIKV-reactive B cells and isolate potent neutralizing monoclonal antibodies (mAbs) with no cross-reactivity to DENV. We group these mAbs into four distinct antigenic groups targeting ZIKV-specific cross-protomer epitopes on the envelope glycoprotein. Co-crystal structures of representative mAbs in complex with ZIKV envelope glycoprotein reveal envelope-dimer epitope and unique dimer-dimer epitope targeting. All four specificities are serologically identified in convalescent humans following ZIKV infection, and representative mAbs from all four groups protect against ZIKV replication in mice. These results provide key insights into ZIKV-specific antigenicity and have implications for ZIKV vaccine, diagnostic, and therapeutic development.
In brief
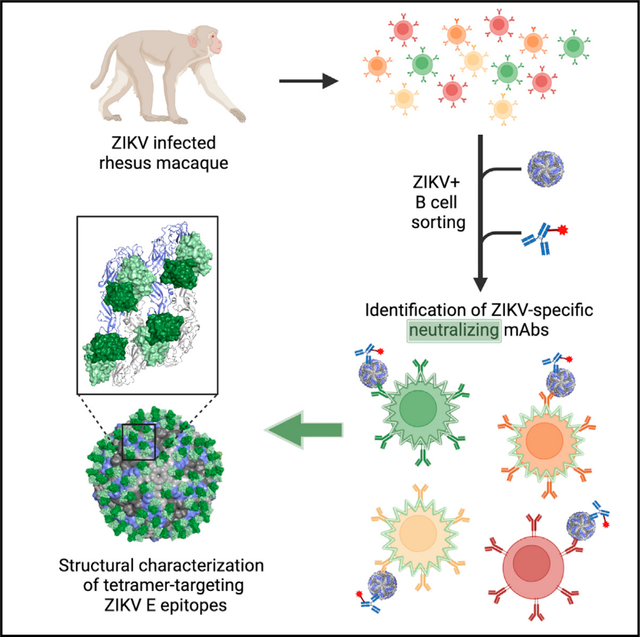
Sankhala et al. use Zika virus as a bait to identify potent neutralizing monoclonal antibodies (mAbs) that are highly specific to Zika virus. These mAbs bind conformational epitopes across the virion dimer-dimer interface and provide protection against infection in a mouse Zika virus challenge model.
Graphical Abstract
INTRODUCTION
Zika virus (ZIKV) is a pathogen that causes significant neurologic and developmental pathology and disease in developing fetuses1 and, in some cases, Guillain-Barré syndrome.2–4 ZIKV was first isolated from a sentinel rhesus monkey in Uganda in 1947 and subsequently described in 1954 in humans.5,6 ZIKV is part of the Flaviviridae family and is transmitted by Aedes aegypti mosquitoes.7 ZIKV can also be transmitted by sexual contact, in addition to perinatal and blood-borne routes.2,4,8–14 ZIKV co-circulates with dengue virus (DENV) in similar geographic locations, while most ZIKV infections that are detected in the United States occur in flavivirus-naive travelers returning from ZIKV-endemic areas.15,16 The ZIKV and DENV envelope (E) glycoproteins elicit immunologic cross-reactivity due to genetic and structural similarity.17 ZIKV vaccination strategies must avoid antibody-dependent enhancement (ADE) of DENV infection.18–20
A vital step to understanding the specificity of protective antibody responses during ZIKV infection is molecular understanding of ZIKV neutralizing monoclonal antibodies (mAbs); this is critical for the development of antibody therapies and vaccines for ZIKV.21–24 The major target of ZIKV-specific mAbs is the viral envelope (E) glycoprotein, which mediates virus attachment, entry, and fusion with endosomal membranes of host cells.25,26 The monomeric E glycoprotein consists of three domains (DI, DII, and DIII), which, via cross-protomer interactions, form a sophisticated icosahedral symmetry on the virus surface.27 Critically, ZIKV neutralizing antibodies target conformational and quaternary epitopes that require higher-order structures on intact virions not fully available on individual E protomers.28,29
Most prior studies that have structurally characterized the epitopes of mAbs against ZIKV have focused on individuals with prior flavivirus exposure.24,29–32 This approach is complicated by B cells from pre-existing DENV immune responses.33 As such, less is known about the immune responses to the ZIKV E glycoprotein targeting in truly flavivirus-naive individuals. Analyses of the B cell antibody repertoire in a flavivirus-naive ZIKV-infected human indicated that more than 60% of B cell responses targeted unknown regions of the E glycoprotein.29,31
We have shown that flavivirus-naive rhesus macaques infected with ZIKV developed high titers of neutralizing antibodies.34 To characterize the antibody repertoire elicited in a naive rhesus macaque following primary ZIKV infection, we isolated neutralizing antibodies using an atypical B cell sorting strategy employing whole ZIKV virions as sorting probes and performed immunological, biochemical, and structural characterization of these mAbs. In addition, we assessed the ability of these mAbs to protect mice against ZIKV infection, and the prevalence of antibodies targeting similar epitopes during ZIKV infection was determined. Overall, these results revealed multiple mechanisms of immune targeting specific to ZIKV to enable enhanced antibody efficacy and vaccine safety development efforts.
RESULTS
Isolation of ZIKV-reactive antibodies from a flavivirus-naive ZIKV-infected macaque
We and others previously demonstrated that flavivirus-naive macaques developed high neutralizing antibody titers following ZIKV infection.33,34 To capture as many ZIKV-reactive B cell specificities as possible, and particularly those recognizing quaternary neutralizing epitopes, we developed a sequential cell-staining strategy based on whole ZIKV virions (Figures 1A and 1B). Unlabeled ZIKV virions were incubated with peripheral blood mononuclear cells (PBMCs) from a flavivirus-naive ZIKV-infected rhesus macaque (10U032) at day 14 post-infection when high titers of neutralization were observed.34 ZIKV-reactive activated CD19+ B cells and plasmablasts (CD19+ CD38+, CXCR5hi/lo) were identified by secondary staining using fluorochrome-labeled mAb 4G2, a fusion-loop-targeting mAb (Figures 1A and 1B).35 Antibody heavy- and light-chain gene segments were amplified from B cells using nested RTP-CR36–38 and sequenced. Immunoglobulin (Ig) G and IgM-targeted PCR was used to generate a total of 40 matched heavy-light chain pairs.
Figure 1. Isolation of ZIKV-reactive antibodies from a flavivirus-naive ZIKV-infected macaque.
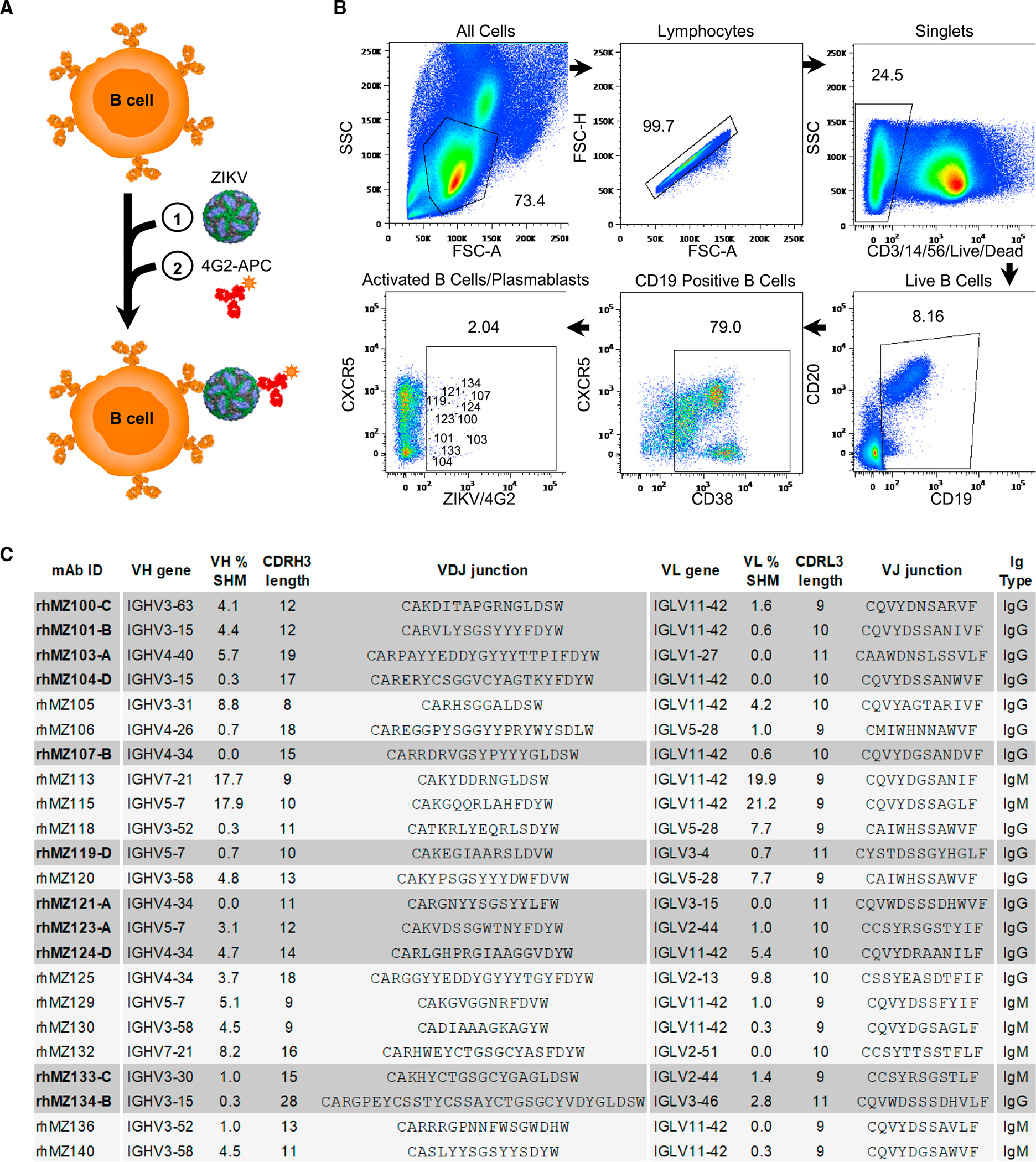
(A) Schematic of the sequential staining strategy of ZIKV-specific B cells. (1) Incubation of PBMCs with unlabeled whole ZIKV virions followed by (2) incubation with fusion-loop-targeting antibody 4G2-APC conjugate.
(B) Isolation of ZIKV-reactive activated B cells and plasmablasts from rhesus macaque 10U032 peripheral blood at 14 days post infection.34 Flow cytometry gates show the percentage of cells identified for each phenotypic population. CD19+CD38+CXCR5hi/lo4G2+ ZIKV-specific B cells sorted and sequenced are indicated in the last gate, with cells encoding neutralizing antibodies labeled with the matching rhMZ antibody number.
(C) Genetic, binding, and neutralization characteristics of isolated ZIKV-reactive mAbs. Antibody nomenclature is in an abbreviated form that indicates species, source, viral probe, clone, and antigenic group. V(D)J assignments were performed using IgBLAST. Antibodies positive in the ZIKV (PRVABC59) MN screen are highlighted in gray.
See also Figures S1, S3, and Tables S1 and S2.
All mAbs were expressed as rhesus macaque IgG1 molecules and screened for ZIKV neutralization and binding to ZIKV soluble E protein (ZIKV E). Twenty-three mAbs bound to ZIKV E (Figures 1C and S1A). Sequence analysis of the antibodies revealed that these 23 ZIKV E-reactive antibodies belonged to 19 independent clonal families that displayed low levels (<10%) of somatic hypermutation (SHM) (Figure 1C; Table S2). Antibodies with low SHM isolated from ZIKV-reactive B cells were also previously reported from flavivirus-naive humans23,29,31 and in other primary infections.39 Two mAbs demonstrated high SHM levels of more than 15% (Figure 1C), with the caveat that this high level may be because of incorrect V gene assignment in IgBlast, due to low coverage within the rhesus database.40 ZIKV E-reactive antibodies showed great diversity in the Heavy Variable (VH) gene usage and complementarity-determining region (CDR) H3 length, ranging from 10 to 30 residues (Figure 1C). ZIKV-specific antibody Light Variable (VL) gene usage was strongly biased toward VL11.42 (50% of the clones).
Antibody characteristics define four ZIKV-specific antigenic groups
Screening in a micro-neutralization (MN) assay using Vero cell culture-produced ZIKV revealed that 11 mAbs were capable of ZIKV neutralization (Figures 1C, 2A, and S1A). The majority (90%) of the neutralizing antibodies were derived from IgG-expressing B cells, with only one IgM-derived neutralizing antibody, rhMZ133-C, (abbreviated for rhesus MHRP ZIKV 133 group C) (Figures 1C, 2A, and 2B). We characterized the 11 neutralizing mAbs to determine specific 50% inhibitory concentration values (IC50; Figures 2A and 2B). While most neutralizing mAbs displayed modest MN activity against a Puerto Rico ZIKV strain (PRVABC59) with IC50 in the μg/mL range, two mAbs, rhMZ133-C and rhMZ134-B, demonstrated greater potencies, similar to or surpassing the EDE1-C8 reference antibody41,42 (Figures 2A and 2B). Remarkably, rhMZ134-B had an IC50 of 5.8 ng/mL, similar to that of Z004, one of the most potent ZIKV neutralizing antibodies reported to date24,43 (Figures 2A and 2B). Although neutralizing mAbs demonstrated significantly higher binding responses than non-neutralizers in the Biolayer Interferometry (BLI) assay (p = 0.0005, Mann-Whitney t test), the magnitude of binding did not correlate with neutralization (Figures 2, S1C, and S1D). To confirm the observed neutralization properties in an assay that would better reflect initial ZIKV infection events, we employed a flow cytometry-based assay (FlowNT50) measuring single-cell infection of human monocytes by a Brazilian strain of ZIKV (Paraiba_01) produced in mosquito cells (Figure 2B). The antibodies typically performed similarly in the FlowNT50 assay and the MN assay. rhMZ134-B showed the highest neutralization potency (IC50 of 15 ng/mL) followed by rhMZ119-D (IC50 of 36 ng/mL), while rhMZ133-C did not perform as well (IC50 ≈ 1 μg/mL) in the FlowNT50 neutralization assay (Figures 2B and S2A). To compare these mAbs to others previously described, a third neutralization assay was performed using the plaque reduction neutralization test (PRNT) with ZIKV (Paraiba_01), assessing five rhMZ antibodies with the highest neutralizing potency as identified by MN or FlowNT50 neutralization assays (Figure S2). The results showed a similar trend compared to the other neutralization assays, with rhMZ134-B potency (IC50 of 30 ng/mL) again being similar to Z004 (IC50 of 20 ng/mL) (Figure S2B). In addition, rhMZ134-B demonstrated broad PRNT neutralization against Ugandan and Thai strains with IC50 of 50 and 20 ng/mL, respectively (Figure S2C). Finally, to investigate whether the isolated antibodies recognized ZIKV-specific or flavivirus cross-reactive epitopes, we performed a neutralization screen against a panel of seven flaviviruses, including all four DENV serotypes, Japanese encephalitis virus (JEV), West Nile virus (WNV) and yellow fever virus (YFV). None of the mAbs displayed neutralization activity even when using a high antibody concentration of 100 μ/mL, indicating that all mAbs were specific for ZIKV (Figure 2B). Consistent with these results, the neutralizing mAbs did not bind a panel of flavivirus E proteins, including DENV1–4, JEV and YFV, confirming their specificity for ZIKV (Figure S1B).
Figure 2. Neutralizing and binding characteristics define four ZIKV-specific epitopes targeted on the E glycoprotein.
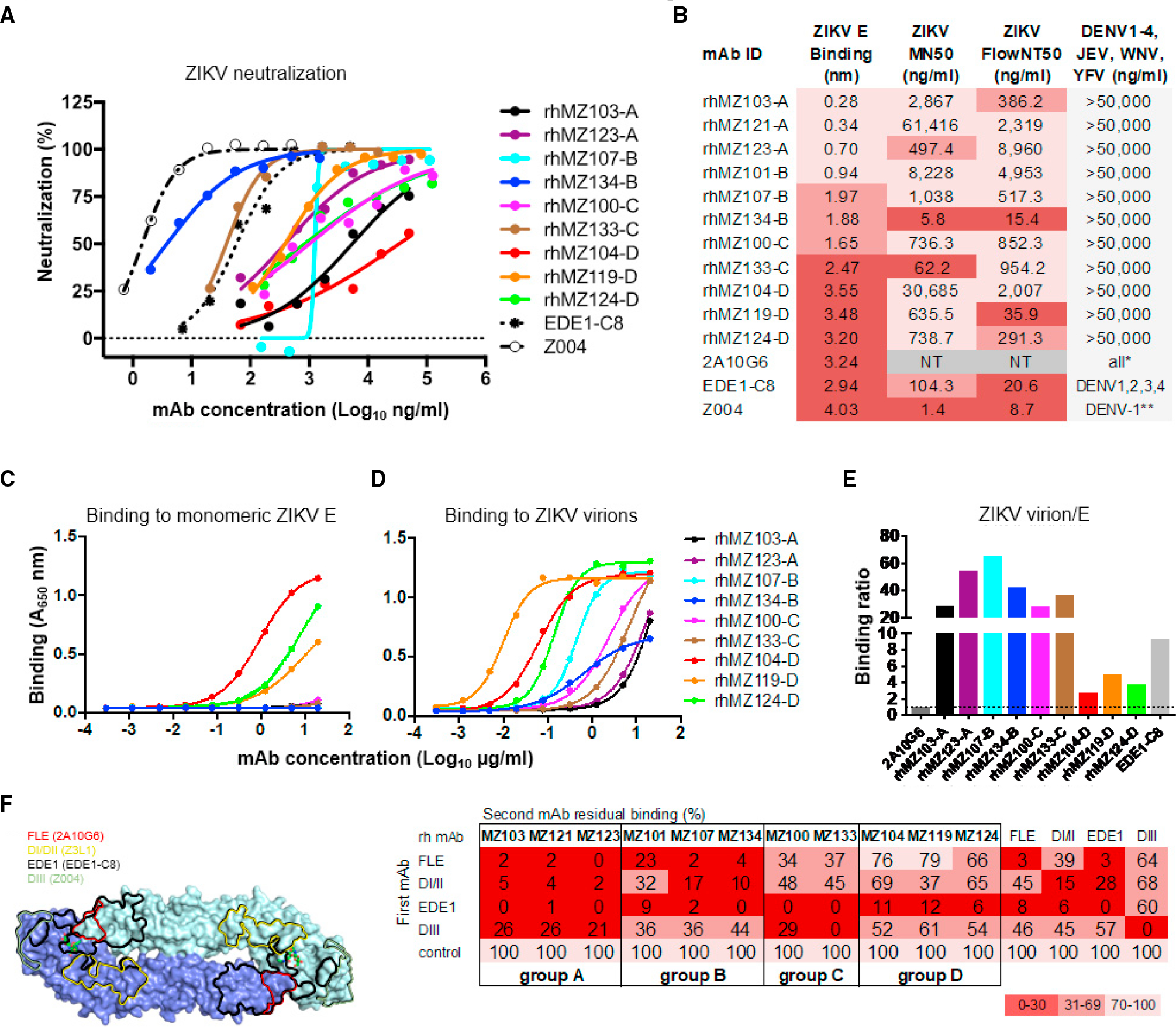
(A) Neutralization against ZIKV PRVABC59 was assessed by MN assay in Vero cells. Shown are neutralization curves compared to the EDE1-C8 and Z004 controls.
(B) Summary of mAb binding and neutralization activities. Antibodies were screened for binding to recombinant ZIKV E by BLI. Values indicate mean binding responses calculated from two independent experiments. Neutralization IC50 values (ng/mL) are shown for MN, and flow-based assay (FlowNT) in Vero, and U937-DC-SIGN cells, respectively. Cross-neutralization screen of a panel of seven flaviviruses (DENV1–4, JEV, WNV, and YFV). Shading represents binding or neutralization strength ranging from strong (dark) to weak (light); NT, not tested.
(C and D) mAb binding of monomeric ZIKV sE (C) and ZIKV (D) by ELISA. Antibodies were titrated using 4-fold dilution series starting from 20 μg/mL. Values indicate mean binding responses calculated from two independent experiments.
(E) Relative ratio of binding to Zika virions compared to E indicating quaternary targeting (calculated from C and D). To directly compare binding activities between the isolated macaque mAbs and human mAb controls, we calculated whole-virus/E binding ratios by using A450 values obtained at 20 μg/mL with their respective secondary antibodies. Binding ratio of 2A10G6, an FLE-directed antibody, was arbitrarily set at 1 (dotted line) as this antibody binds equally well to both monomeric E and ZIKV virion. Antibodies with ratio values >1 indicated preferential targeting of quaternary epitopes (such as EDE), whereas ratios at or below 1 were characteristic of monomeric recognition similar to FLE antibodies.
(F) Binding competition with control antibodies defined four targeting epitope groups. Left: control antibody epitopes mapped on the ZIKV E dimer structure. Right: four distinct antibody competition profiles were identified in a BLI-based competition assay. Values represent the percentage of residual binding of the indicated second antibody after prior saturation of ZIKV E with the indicated first antibody. Shading from dark to light indicates competition strength ranging from strong (0%–30%), to intermediate (31%–69%), to weak/none (70%–100%). The negative control mAb was an HIV-specific mAb VRC01 that has no reactivity to ZIKV E.
See also Figures S1 and S2 and Tables S2 and S3.
To understand the epitopes targeted by this set of ZIKV-specific neutralizing mAbs, we assessed binding to monomeric ZIKV E and whole ZIKV. Binding to monomeric ZIKV E was performed by ELISA where quaternary antibodies, such as EDE1-C8, are poorly reactive,44 while antibodies to the monomeric E protein, such as the fusion-loop epitope (FLE)-directed mAb 2A10G6, bind robustly (Figure S2D).45 In contrast, since utilizing whole ZIKV virions in a virus-capture ELISA assay allows for the display of monomeric and quaternary epitopes, both EDE1-C8 and 2A10G6 showed strong binding to ZIKV (Figure S2D). Interestingly, binding to whole ZIKV virion mirrored the responses to ZIKV E observed in the BLI assay (Figure S1A), suggesting that both monomeric and E dimer epitopes (EDEs) are available. The majority of the ZIKV-specific mAbs did not demonstrate binding to monomeric ZIKV E in ELISA (Figure 2C). Some reactivity was observed for rhMZ104-D, rhMZ119-D, and rhMZ124-D, which were also identified as the strongest binders to ZIKV E by BLI (Figures 2B and 2C). All ZIKV-specific neutralizing mAbs bound strongly to ZIKV virions (Figure 2D). To determine the quaternary specificity of the neutralizing antibodies, we calculated the ZIKV virion/ZIKV E binding ratios relative to the FLE control antibody (2A10G6); a ratio of 1 reflects an equally strong affinity for monomeric E and ZIKV virion (Figure 2E). In contrast, the binding of the EDE1-C8 control, with a ratio of approximately 10, was strongly biased toward whole ZIKV virion binding, consistent with its quaternary epitope specificity. Intermediate binding ratios between 2 and 5 were obtained for group D mAbs, rhMZ104-D, rhMZ119-D, and rhMZ124-D, indicating that they partially bound to monomeric ZIKV E but have significant quaternary affinity. Neutralizing antibodies from groups A–C had binding ratios over 20 (Figure 2E), indicating preference for quaternary epitopes found only on ZIKV virions. Intrigued by potent neutralizing activities of these mAbs, we next tested their binding affinities for the ZIKV E glycoprotein. Binding affinities for group D antibodies were on par with EDE1-C8 and slightly less for group B and C mAbs (Table S3).
We performed binding competition experiments to delineate the antigenic sites (Figure 2F). Control mAbs against previously identified sites of vulnerability such as the FLE (mAb 2A10G6), DI/II (mAb Z3L145), the EDE1 (mAb EDE1-C8), and DIII (mAb Z004) were used to assess binding to ZIKV E using BLI. When the 11 ZIKV-specific macaque neutralizing mAbs were used in these competition assays, their pattern of competition fell into four groups with overlapping but discrete features (Figure 2F). Antibodies within group A were competed by all four control mAbs to varying degrees. Group B antibodies resembled the competition profile obtained with the EDE1 control antibody, suggesting that they might represent unique ZIKV-specific EDE antibodies. Group C was competed by EDE1 and DIII mAbs, whereas group D mAbs were only sensitive to EDE1 mAb competition. Group C and D antibodies were of particular interest as their distinctive competition profiles indicated that they might target previously uncharacterized neutralizing epitopes.
Crystal structure of ZIKV-specific EDE antibody rhMZ107-B Fab in complex with ZIKV E glycoprotein
To understand the structural recognition of the ZIKV-specific EDE-like antibodies, crystal structures of representative mAbs from each group were determined alone and in complex with ZIKV E. Group A mAbs did not form stable complexes with ZIKV E and failed to crystallize. However, we were able to crystallize the antigen-binding fragment (Fab) rhMZ103-A alone and determine the structure to a resolution of 1.87 Å (Figures S3A and S3B; Table S4A). From antigenic specificity group B, we determined the crystal structure of rhMZ107-B alone (2.1 Å resolution) and in complex with ZIKV E glycoprotein at 3.2 Å resolution (Figures 3A and S3C; Tables S4A, S4B, S5A, and S5B). Within the asymmetric unit, four rhMZ107-B Fab variable region (Fv) and four ZIKV E glycoproteins were observed, with each Fv binding an epitope that spans three protomers (I–III) with protomer contact regions of 295.3, 852.8, and 64.9 Å2 buried surface area (BSA), respectively (Table S5B). The largest antibody contact area was on the DII of protomer II, followed by recognition of the DIII of protomer I, adjacent to the glycan-154, with minor contacts on the DI of a third protomer (III) (Figures 3A and 3B; Table S5B). The antibody light chain had more contacts with ZIKV E protomers compared to the heavy chain (483.4 Å2 heavy chain; 729.6 Å2 light chain). mAb rhMZ107-B recognition of ZIKV E involved almost exclusively germline-encoded residues except for two residues encoded through recombination. In the light chain, a single CDR L3 residue glycine 93, which makes main chain contact with ZIKV E, was mutated from the VL11.42 gene-encoded residue serine (Figure S3A). Modeling antibody recognition in the context of the mature ZIKV (PDB: 5IRE), the rhMZ107-B epitope is fully accessible and all 180 protomer epitopes are available without any E glycoprotein structural rearrangement needed (Figure S4D).
Figure 3. Crystal structure of ZIKV-specific EDE antibody rhMZ107-B in complex with ZIKV E glycoprotein.
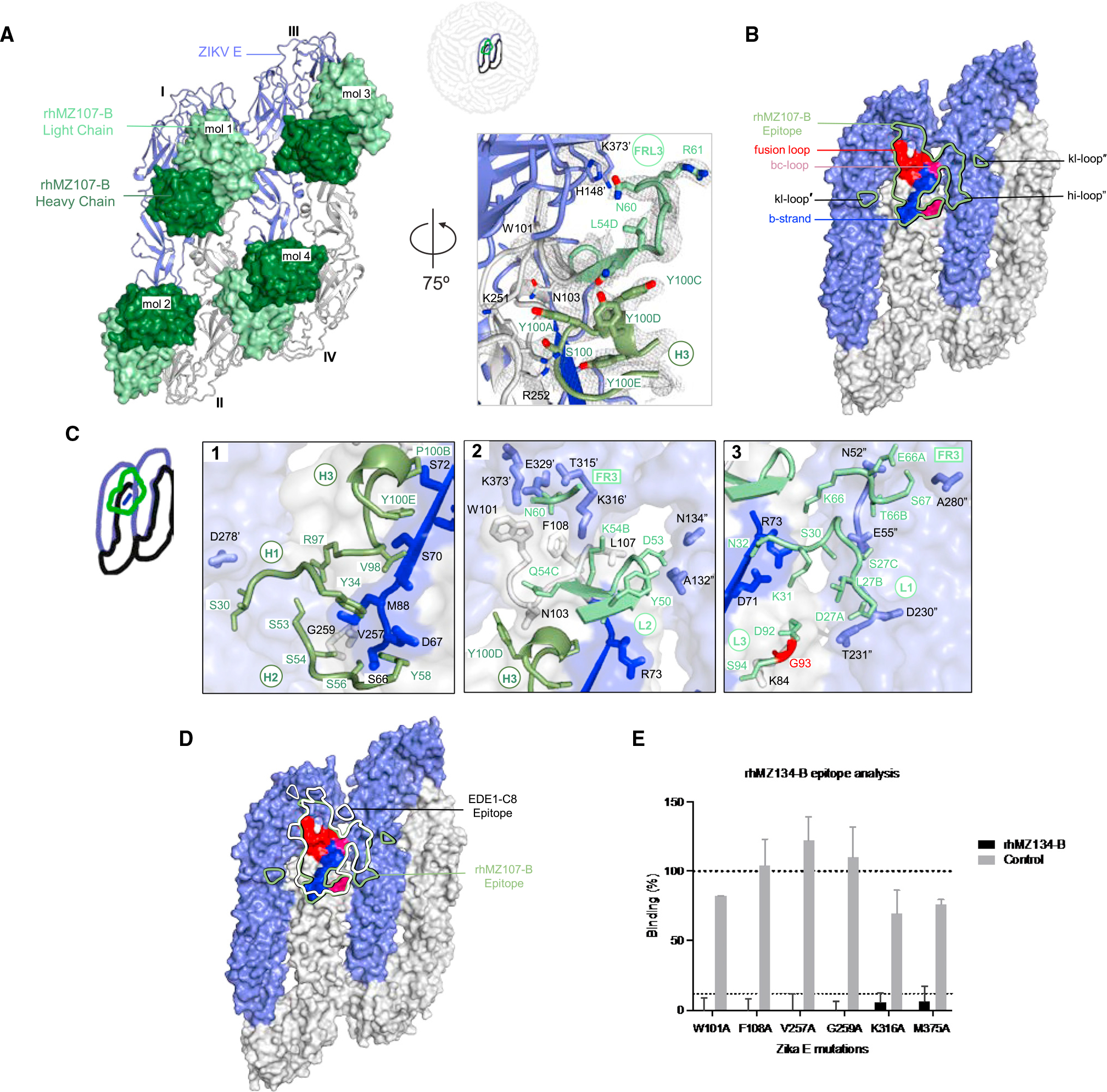
(A) Left: top view of the co-crystal structure of rhMZ107-B in complex with ZIKV E (PRABC59). rhMZ107-B Fv heavy and light chains are shown in surface representation and are colored dark and light green, respectively, while four ZIKV E protomers are shown in ribbon representation and colored blue and gray. Four ZIKV E protomers, left to right, are labeled as I–IV. Right: 2Fo-Fc electron density for the rhMZ107-B (mol 1) and ZIKV E interface residues is shown as gray mesh (contoured at 1.5σ).
(B) Epitope footprint of rhMZ107-B antibody (mol 1) is shown as solid green line, displayed on four ZIKV E protomers shown in surface representation. TherhMZ107-B epitope extends across protomers I–III. Relevant antigenic ZIKV E regions within the epitope are labeled (fusion loop is in red, b strand in blue, bc-loop in magenta, and other loop regions are labeled). Antigenic regions of protomers I and III are marked with prime (′) and double prime (″), respectively.
(C) rhMZ107-B contact residues are shown as sticks based on (1) CDRs H1, H2, and H3; (2) CDR H3 and L2; (3) CDR L1, L3, and FR L3 antibody-contacting regions. ZIKV E contact residues of protomers I and III are marked with prime (′) and double prime (″), respectively. The b strand residues 63–73 on protomer II are colored dark blue. mAb somatic hypermutation (SHM) residues are colored bright red.
(D) Epitopes for rhMZ107-B and EDE1-C8 are represented with green and white lines, respectively. EDE1-C8 (PDB: 5LBS) antibody was overlaid onto therhMZ107-B ZIKV E structure to map the epitopes.
(E) Shotgun mutagenesis ZIKV E epitope analysis for rhMZ134-B antibody. Relative binding to ZIKV prM/E for individual mutations is plotted. Residues from which substitution to alanine causes >90% loss in binding were considered important for binding; limit of detection: 10% (gray dotted line). Error bars indicate mean ± SEM from two independent experiments.
See also Figures S3–S5 and Tables S4 and S5.
rhMZ107-B recognized the DII of ZIKV E utilizing CDRs H1–H3 and L1–L3 focused on the b strand in the center of the epitope (residues 65–74). Additional recognition of DIII from an adjacent protomer I occurred through the light chain CDR L2 and framework 3 (FR3), while recognition of the DI from the third protomer (III) was facilitated by CDR L1, L2, and FR3 (Figure 3C). Features of the rhMZ107-B antibody recognition are reminiscent of EDE1-C8 recognition,42 which had reactivity against ZIKV, as well as multiple strains of DENV (Figure 2B). In contrast to EDE1-C8, rhMZ group B mAbs inclusive of rhMZ107-B were ZIKV specific (Figure 2B). Comparison of the epitope-contact residues of rhMZ107-B and EDE1-C8 showed similar contacts (Figures 3D and S5C; Table S5) with a few notable sequence differences. rhMZ107-B makes close contact with the DI-glycan loop (BSA 74.0 Å2) and DE-loop (BSA 83.0 Å2) on one protomer and the b strand (residues 65–74; BSA 400.0 Å2) on the neighboring E protomer (Figure 3A). The ZIKV b strand, has no sequence similarity with the b strand on DENV E. Moreover, DENV E glycoprotein is glycosylated at residue N67 within the b strand, while this glycosylation site is absent in ZIKV E. Both the DI-glycan loop and the DE-loop in the DENV E glycoprotein are shorter with significant sequence differences (68% and 80% sequence similarity, respectively). Overall, these differences in key contact residues on the E glycoproteins between the two viruses explains the ZIKV specificity. Although crystal structures were not solved for the group B rhMZ134-B—the most potent ZIKV-specific mAb isolated—we screened rhMZ134-B for recognition of ZIKV E glycoprotein by shotgun alanine/serine-scanning mutagenesis. Residues W101, F108, V257, G259, K316, and M375 were highlighted as contact residues, similar to the rhMZ107-B epitope (Figure 3E) and other EDE1-like antibodies.42,46 The disparity in neutralization potency (μg/mL range for rhMZ134-B and EDE1-C8, and mg/mL range for rhMZ107-B) between these antibodies is likely due to slight differences in epitope recognition in the viral context also indicated by binding differences (Figure 2D). These large potency differences between antibodies targeting overlapping epitopes have been previously observed for multiple viral neutralizing antibodies.43,47–49
Crystal structure of inter-dimer-epitope antibody rhMZ100-C Fab in complex with ZIKV E glycoprotein
To understand the structural basis for the group C antigenic specificity, we determined the crystal structure of rhMZ100-C alone (2.1 Å resolution) and in complex with ZIKV E glycoprotein at 2.8 Å resolution (Figures 4 and S3D; Tables S4A and S4B). Within the asymmetric unit, two rhMZ100-C Fab molecules and four ZIKV E glycoprotein protomers (two dimers) were observed with both the dimers flipped at an angle of 180° with respect to each other (Figure S4A). Within a dimer, each Fab bound to a single protomer with a contact region of 380.8 Å2 BSA through the light chain and 138.9 Å2 from the heavy chain that focused on DII. rhMZ100-C recognized the DII of ZIKV E utilizing CDR L1–3 and heavy-chain CDR H3, centered on the ZIKV E b strand (Figures 4B and 4C). Antibody rhMZ100-C used 95% heavy-chain germline-encoded residues and 97% in the light chain (Figure S3A). We modeled rhMZ100-C binding in the context of the mature ZIKV (PDB: 5IRE) and identified additional significant contacts in the DI/DII of an adjacent ZIKV E protomer that would increase the light- and heavy-chain contact region to 544.0 and 975.0 Å2, respectively (Figure S4A). In this model, rhMZ100-C recognized two molecules of ZIKV E (993.2 and 526.4 Å2 BSA) at the inter-dimer interface in a ZIKV E-tetramer with only 60 binding sites accessible in the context of the mature ZIKV, since this epitope spanned the center of two dimers, and thus only half of the epitope was available on the “end” dimers that form the raft-raft interface (Figure S4D). The rhMZ100-C epitope has significant sequence difference in the DENV-2 E (Figures 6A and S5D). Features of the rhMZ100-C antibody recognition of ZIKV were like ZIKV-117 mAb,50,51 which is highly potent and ZIKV specific (Figure 4D). However, ZIKV-117 recognizes an inter-dimer epitope that is less centered across the inter-dimer interface, with predominant recognition of one protomer (total BSA, 263.9 Å2) which is distinctly different to the rhMZ100-C epitope (Figure 4E).
Figure 4. Crystal structure of inter-dimer-epitope antibody rhMZ100-C Fab in complex with ZIKV E glycoprotein.
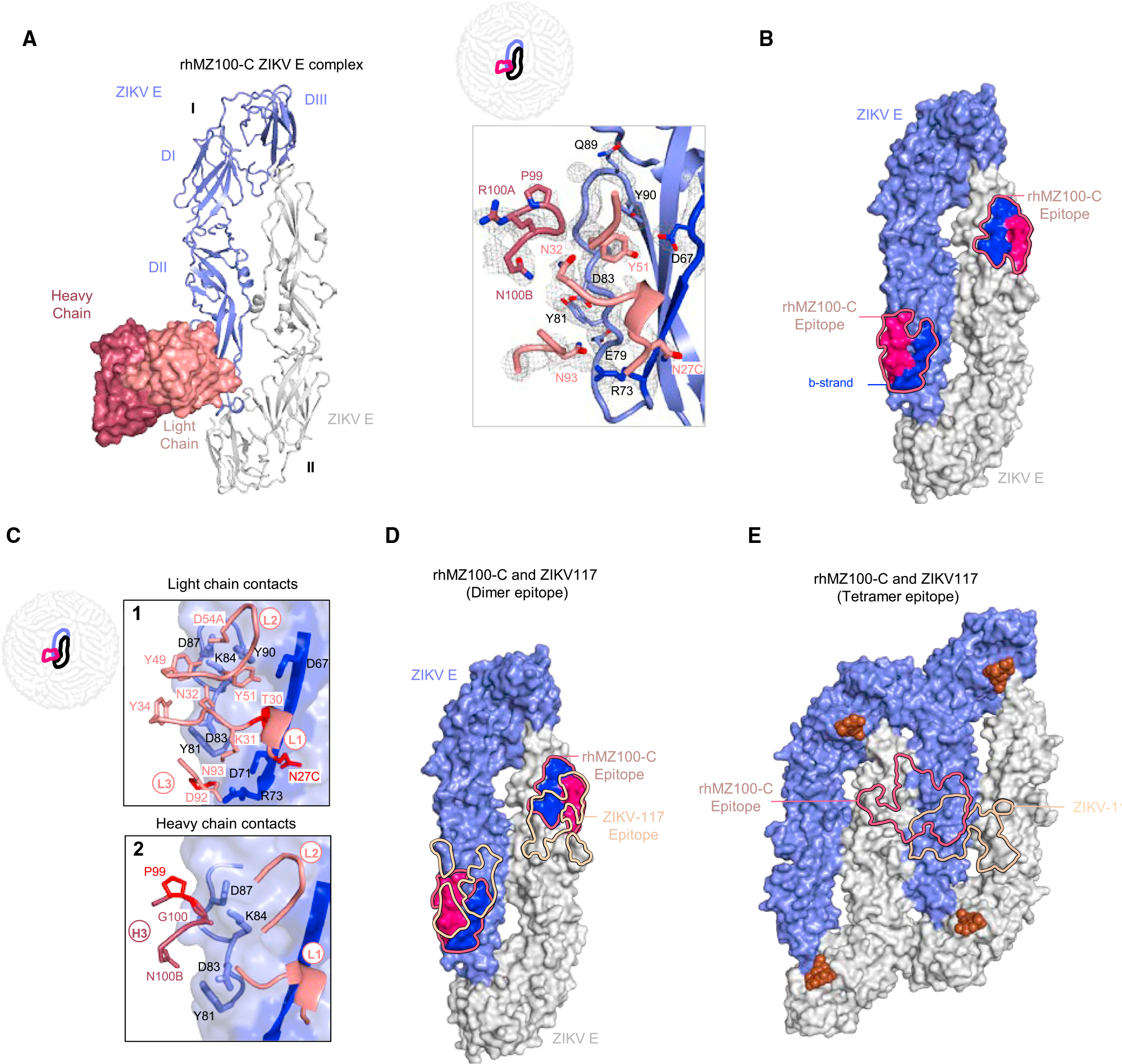
(A) Left: top view of the co-crystal structure of rhMZ100-C in complex with ZIKV E (PRABC59). rhMZ100-C Fv heavy and light chains are colored dark and light raspberry, respectively, and are shown in surface representation, while two ZIKV E protomers are shown in ribbon representation colored blue and gray. ZIKV E protomers, left to right, are labeled as I and II. Right: 2Fo-Fc electron density for the rhMZ100-C and ZIKV E interface residues is shown as gray mesh (contoured at 1.5σ).
(B) Epitope footprint of rhMZ100-C antibody is indicated with a solid raspberry-colored line displayed on two ZIKV E protomers (surface representation). Relevant antigenic ZIKV E regions within the epitope are labeled (b strand is shown in blue and rest of the rhMZ100 epitope is shown in raspberry color).
(C) rhMZ100-C contact residues are shown as sticks based on (1) CDRs L1, L2, and L3; (2) CDR H3 antibody-contacting regions; b strand residues 63–73 on protomer I are highlighted in dark blue color. SHM residues are colored bright red.
(D) Epitopes for rhMZ100-C and ZIKV-117 antibodies are represented with raspberry and teal colored lines, respectively. ZIKV-117 (PDB: 5UHY) antibody was overlaid onto the rhMZ100-C ZIKV E structure to map the epitope.
(E) rhMZ100-C and ZIKV-117 epitopes are mapped onto a ZIKV E-tetramer (PDB: 5IRE).
See also Figures S3–S5 and Tables S4 and S5.
Figure 6. Comparison of DENV and ZIKV antibody specificity.
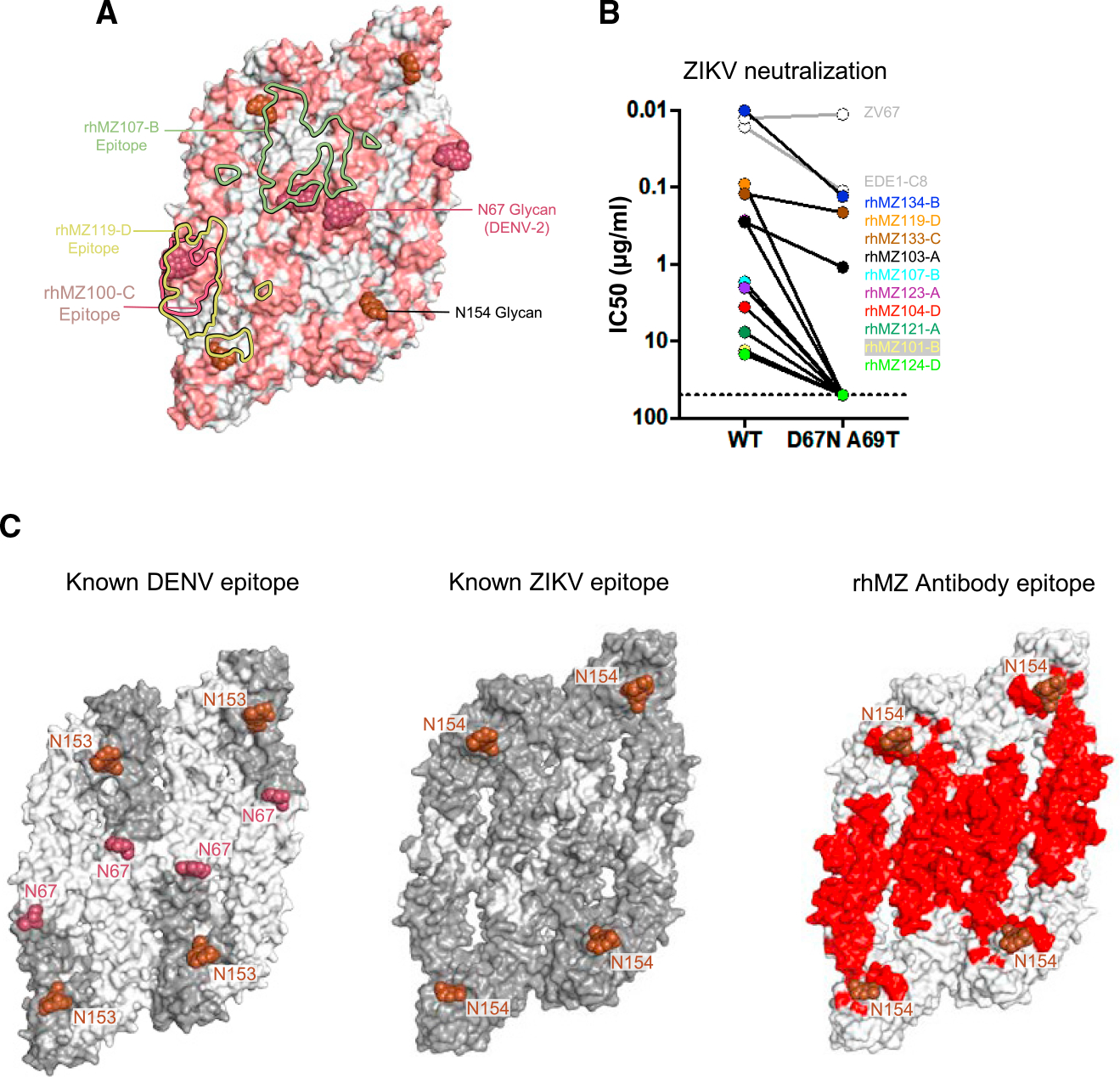
(A) Sequence differences between ZIKV E and DENV-2 E are mapped onto the ZIKV structure (PDB: 5IRE). Sequence and positional differences between ZIKV and DENV-2 are colored light red, while identical residues are colored white. Glycan-154 and DENV glycan-67 are shown in sphere representation colored brown and raspberry, respectively. The antibody binding footprints of rhMZ107-B, rhMZ100-C, and rhMZ119-D antibodies are shown as green, raspberry, and yellow solid lines, respectively.
(B) Neutralization (IC50, mg/mL) of wild-type (WT) and D67N-A69T mutant ZIKV performed in the ZIKV/H/PF2013 background using a reporter virus particle (RVP) assay. The addition of glycan-67 to ZIKV interfered with epitope recognition and abrogated or eliminated neutralization.
(C) Epitope mapping of structurally defined antibodies mapped onto four protomers of DENV (left) and ZIKV (middle). Residues contacted by previously described mAbs are colored dark gray, and residues not previously identified prior to this study are indicated in white. Only previously identified mAb structures with resolution greater than 4 Å were used since the contact residues are clearly interpretable. Newly identified residues contacted by rhMZ mAbs described in this study are colored red (right). Glycan sites at positions 153 or 154 are indicated in rose color.
See also Figures S5 and S7 and Table S6.
Structure of inter-dimer-epitope antibodies rhMZ119-D and rhMZ104-D in complex with ZIKV E glycoprotein
To understand the antigenic specificity group D recognition, we determined the crystal structures of rhMZ119-D and rhMZ104-D Fabs alone and in complex with ZIKV E glycoprotein (Figures 5, S3E, S3F, S4B, and S4C). Crystal structures of rhMZ119-D and rhMZ104-D complexes were determined at 3.58 and 2.80 Å resolutions, respectively (Figure 5; Tables S4A and S4B). While both antibodies approach ZIKV E glycoprotein from two different angles, the antibody-antigen interactions are primarily centered on DII domain (Figures 5A, 5C, and 5G). For rhMZ119-D complex, only one molecule of antibody and antigen were observed within an asymmetric unit (Figure S4C), and re-construction of a second ZIKV E molecule from crystal contacts suggests additional contacting residues on DI and DII domains of a neighboring protomer (Figures 5A and 5B). Total BSA for rhMZ119-D/ZIKV E interaction is 1,275.0 Å2 and heavy and light chains contribute 506.3 and 768.5 Å2, respectively. The antibody-antigen interactions are centered on a region consisting of the b strand (residues S64–S72), the bc-loop (residues C74, L82-K84, D87, Q89, and Y90), the fusion loop (residues R99, N103, and G104), d strand (residues C116, K118, A120, and S122), hi-loop (residue T233), and the ij loop (residues K251 and R252), contacted by side chains from all three CDRs in the heavy and light chains (Figure 5E; Tables S5G and S5H). In addition, rhMZ119-D contacts the N-154 glycan loop (residue M151, N154-T156, G157, E159, and T160) of DI and the kl loop (residue D276 and 279) of DII from the adjacent subunit on the opposite side (Figures 5A, 5B, and 5E; Table S5H). rhMZ119-D also interacts with N-154 glycan from the neighboring protomer (BSA: 109.5 Å2; Figures 5A and 5B). Modeling of rhMZ119-D recognition in the context of the mature ZIKV suggested accessibility of all 180 binding sites on a mature virion (Figure S4D).
Figure 5. Structure of inter-dimer-epitope antibodies rhMZ119-D and rhMZ104-D in complex with ZIKV E glycoprotein.
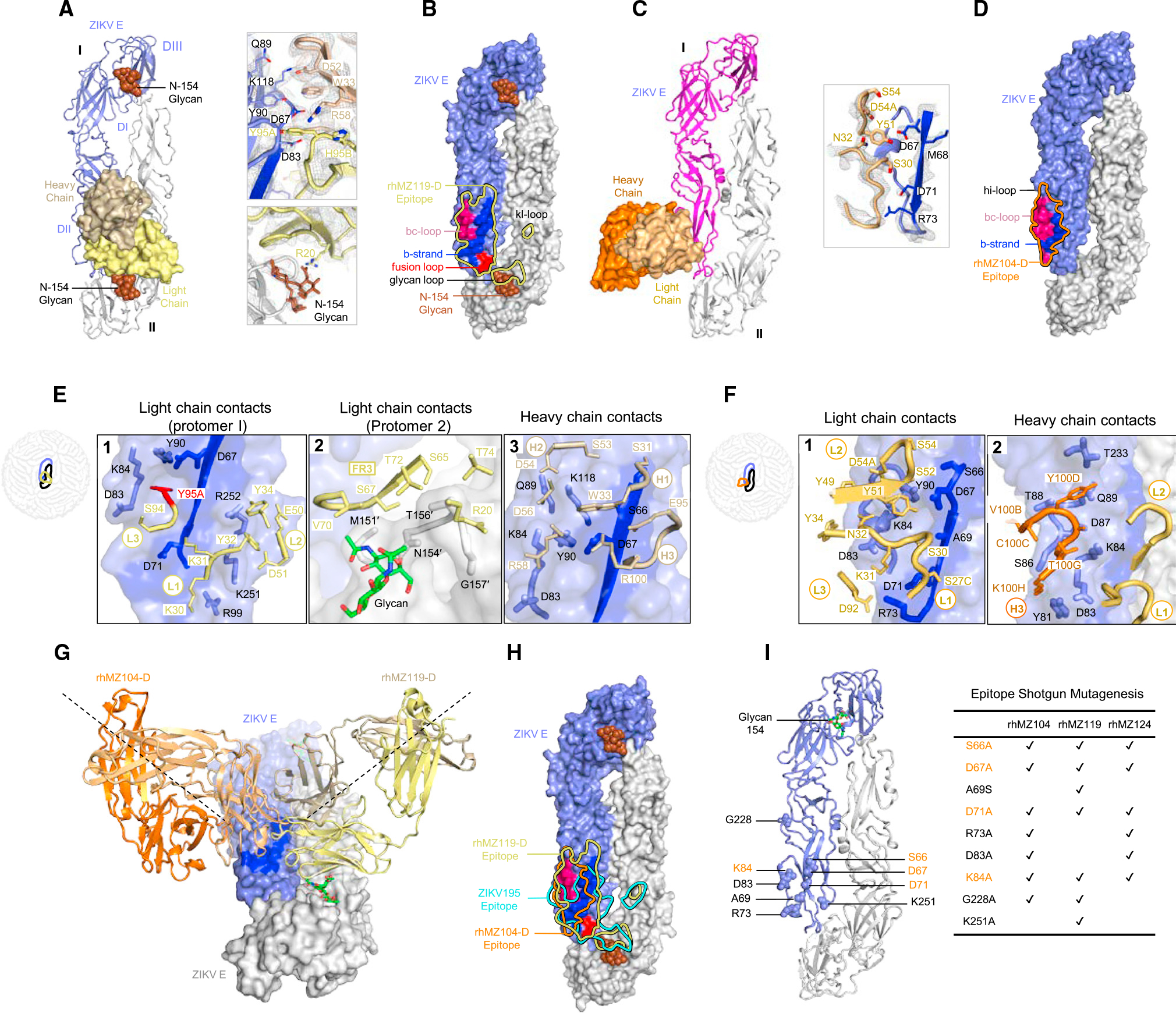
(A) Left: top view of the co-crystal structure of rhMZ119-D in complex with ZIKV E (PRABC59). rhMZ119-D Fab heavy and light chains are shown in surface representation and are colored wheat and yellow, respectively, while two ZIKV E protomers are shown in ribbon representation, colored blue and gray. ZIKV E protomers, left to right, are labeled as I and II. Right: 2Fo-Fc electron density for the rhMZ119-D and ZIKV E interface residues is shown as gray mesh (contoured at 1.5σ).
(B) Epitope footprint of rhMZ119-D antibody is shown as solid yellow colored line, displayed on two ZIKV E protomers in surface representation.
(C) Left: top view of the crystal structure of rhMZ104-D in complex with ZIKV E (PRABC59). rhMZ104-D Fab heavy and light chains are shown in surface representation and are colored with dark and light orange, respectively, while two ZIKV E protomers are shown in ribbon representation and colored blue and gray. ZIKV E protomers, left to right, are labeled as I and II. Right: 2Fo-Fc electron density for the rhMZ104-D and ZIKV E interface residues is shown as gray mesh (contoured at 1.5σ).
(D) Epitope footprint of rhMZ104-D antibody is shown as solid orange colored line, displayed on two ZIKV E protomers in surface representation.
(E) rhMZ119-D contact residues are shown as sticks based on (1) CDRs L1, L2, and L3; (2) CDR FRL3; and (3) CDR H1, H2, and H3 antibody-contacting regions. ZIKV E contact residues of protomer II are marked with prime (′); b strand residues 63–73 on protomer I are highlighted in dark blue color. SHM residues are highlighted in bright red color.
(F) rhMZ104-D contact residues are shown as sticks based on (1) CDRs L1, L2, and L3; (2) CDR H3 antibody-contacting regions; b strand residues 63–73 on protomer I are highlighted in dark blue color.
(G) Antibodies rhMZ119-D and rhMZ104-D are superimposed on ZIKV E protomer I. Both antibodies approach the E dimer at an angle of 90° with respect to each other.
(H) Epitopes for rhMZ104-D, rhMZ119-D, and ZIKV-195 antibodies are represented with orange, yellow, and cyan colored lines, respectively. rhMZ104-D andZIKV-195 (PDB: 6MID) antibodies were overlaid onto the rhMZ119-D ZIKV E structure to map the epitopes.
(I) Group D antibodies were assessed for binding using shotgun mutagenesis epitope mapping. Alanine or serine mutations, which dramatically affected group D antibody binding, are shown in sphere representation on the ZIKV E structure and indicated on the right. Residues important for binding for all group D mAbs are highlighted in orange.
See also Figures S3–S5 and Tables S4 and S5.
The rhMZ104-D complex has one rhMZ104-D Fab, one Fv molecule, and two ZIKV E glycoprotein protomers within the asymmetric unit, with rhMZ104-D binding to the DII region of a single protomer (210.4 Å2 BSA light chain and 385.0 Å2 heavy chain) (Figures 5C, 5D, and S4B; Tables S5E and S5F). Primarily, these contacts are to the b strand (BSA 96.1 Å2) and α helix 1 (BSA 543.0 Å2). rhMZ104-D utilized mainly heavy- and light-chain germline-encoded residues alongside residues in the CDR 3 loops encoded by recombination and somatic mutation (Figure S3A). rhMZ104-D recognized DII utilizing all light-chain CDRs and CDR H3 with the ZIKV E b strand being the major antigen contact region for the light chain, alongside a continuous stretch of residues from position 77–90 that is recognized by both heavy and light chains (Figure 5F). We modeled rhMZ104-D recognition on the ZIKV virion and identified significant additional heavy-chain contacts in the DI/DII of two adjacent ZIKV E protomers increasing the light- and heavy-chain contact region to 678.0 and 2,159.0 Å2, respectively (Figure S4B). Similar to rhMZ100-C, only 60 binding sites were accessible when modeled within the context of the mature ZIKV since the rhMZ104-D epitope spanned the center of two dimers and only half of the epitope was available on the end dimers (Figure S4B). Antibody recognition by the D-epitope group of mAbs were similar to ZIKV-195 mAb,50,51 which is moderately potent against ZIKV with IC50 values in a range of 77–600 ng/mL (Figure 4H). However, there were distinct differences, with the D group of antibodies making more extended contacts with the DII domain residues compared to ZIKV-195 (Figure 4H).
In addition to the structural data describing the rhMZ119-D and rhMZ104-D epitope, we also screened all group D antibodies rhMZ104-D, rhMZ119-D, and rhMZ124-D for common interacting residues and recognition of ZIKV E glycoprotein by shotgun alanine scanning mutagenesis (Figures 5I and S6) to determine common residues. The scanning mutations identified six or seven residues per antibody that significantly altered ZIKV E recognition. In all of the antibodies in this antigenic group, residues S66, D67, D71, and K84 were identified as significant contact residues, and R73, D83, and G228 residue mutations knocked out binding of two of the three group D mAbs (Figures 5I and S6).
Comparison of DENV and ZIKV antibody specificity
Many epitopes targeted by the ZIKV-specific mAbs curiously overlapped with the glycan at position 67 present on DENV E glycoprotein (Figures 6A and S5). Therefore, we assessed the role of the glycan at position 67 on antigenic recognition of ZIKV compared to DENV. In the case of DENV E, this glycan is highly prevalent in virus sequence datasets and has been shown to interact with Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (DC-SIGN).52 We assessed our set of 11 mAbs for neutralization of wild-type ZIKV (H/PF/2013) and a ZIKV E mutant (D67N, A69T) that contains a neoglycan at position 67. For all mAbs, the introduction of the glycan at position 67 resulted in loss of neutralization ranging from a 2-fold reduction for rhMZ133-C, to 10-fold for rhMZ134-B, and complete ablation of neutralization for the remaining eight antibodies, inclusive of all group D mAbs (Figure 6B). The presence of this glycan did not alter DIII-targeting antibodies such as ZV67 but reduced EDE1-C8 neutralization potency. Modeling of rhMZ antibodies in the context of DENV-2 E, which contains a glycan at residue N67, also suggests that all the ZIKV-specific antibodies (rhMZ antibodies and ZIKV-117) clash with N67 glycan, whereas ZIKV-DENV cross-reactive mAbs (EDE1-C8 and EDE2-A11) can bind with minimal clashing (Figures S5A and S5B).
To understand immune responses to ZIKV compared to DENV, all ZIKV- or DENV-related antibody-antigen complex structures (resolution below 6 Å) available in the PDB were analyzed, and epitopes or antibody contact residues were determined (Tables S6A and S6B). These contact residues were mapped onto four ZIKV or DENV protomers (Figure 6C). The vast majority of DENV-targeting antibodies recognize the E glycoprotein DII and DIII regions in areas distal to the N67 glycan site on DENV (Figure 6C; Table S6), which surprisingly leaves a significant region on DENV E that has not been described in antibody-antigen structural studies to date. In contrast, structures of antibody-antigen recognition for ZIKV describing the natural antibody response show that the full surface of the ZIKV E glycoprotein is available (Figure 6C).
Prevalence and protection of ZIKV-specific neutralizing mAbs
We next evaluated the prevalence of similar antibodies in other ZIKV-infected rhesus macaques and humans. We also included plasma from humans with verified DENV experience (DENV+/ZIKV+) to assess whether pre-exposure to DENV would influence the specificity of the antibody responses following infection with ZIKV. We carried out binding competition to the ZIKV E protein, between plasma samples and representative mAbs from each group (Figure S7). Remarkably, binding of group A–D antibodies was almost completely abrogated in the presence of ZIKV E-immune plasma complexes, suggesting that antibodies with similar specificities are commonly elicited during the course of natural ZIKV infection (Figure S7B). Similarly, plasma from DENV/ZIKV-experienced donors efficiently inhibited binding of our ZIKV-specific neutralizing antibodies, indicating that preexposure to DENV had little effect on the generation of these ZIKV-specific responses. Comparable observations were seen from ZIKV-infected rhesus macaques at day 14 post infection (Figure S7B).
Finally, we conducted passive protection experiments in mice to determine whether group-representative neutralizing mAbs would prevent viral replication in vivo. Six neutralizing antibodies of various potencies were infused into groups of naive Balb/c mice (n = 5/group) at a single dose (10 mg/kg). Mice were then challenged with 105 viral particles (102 plaque-forming units) of ZIKV-BR intravenously and viral replication was monitored using RT-PCR53 (Figure 7A). The mAbs identified with the highest potency in neutralization, rhMZ134-B, rhMZ133-C, and rhMZ119-D, conferred total protection from ZIKV replication compared to control mice, where ZIKV viral load peaked at day 3 post challenge (Figure 7B). Partial protection was also observed with the less potent neutralizing antibodies (IC50 within the 1- to 3-μg/mL range in FlowNT50 assays), with one (rhMZ103-A, rhMZ100-C) or two (rhMZ107-B) mice out of five showing delayed but detectable viremia. Next, we assessed the viral spread in the brain, spleen, and lymph node tissues of animals that received rhMZ134-B, rhMZ133-C, and rhMZ119-D mAbs compared to the control group (Figure 7C). While no viral RNA was detected in the brain tissues of any of the groups, saline-infused control mice exhibited moderate to high levels of viral RNA in the spleen and lymph node tissues. Once again, mAbs rhMZ134-B, rhMZ133-C, and rhMZ119-D conferred total protection with no viral RNA in their spleen or lymph nodes.
Figure 7. Protection of ZIKV-specific neutralizing mAbs.
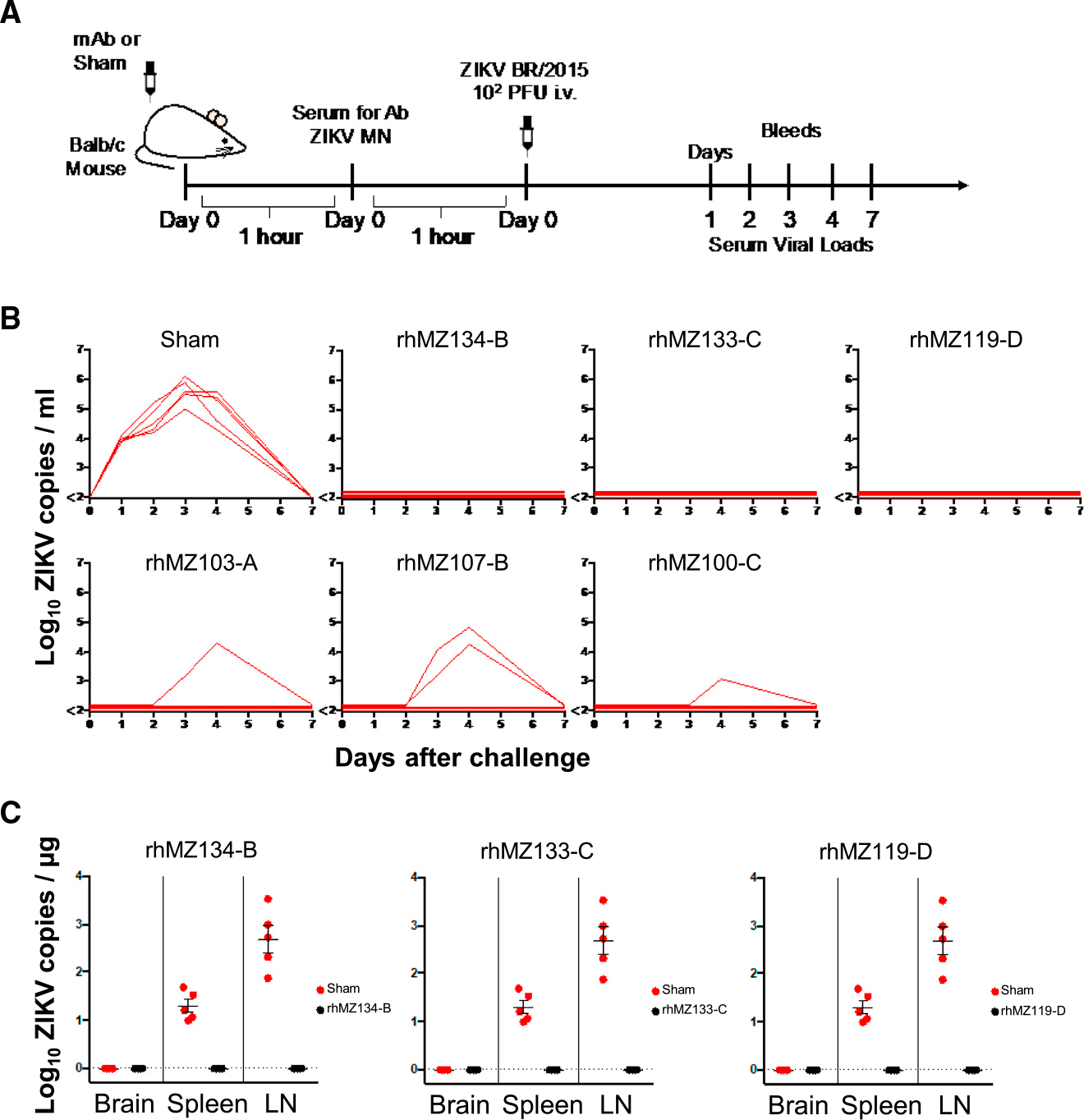
(A) Schematic of passive protection study experimental design. Antibodies were infused intravenously into groups of naive recipient Balb/c mice (n = 5/group) prior to ZIKV-BR challenge. Mice received 200 μg of antibody (10 mg/kg) and were challenged with 105 viral particles (102 plaque-forming units) of ZIKV-BR intravenously 2 h after infusion.
(B) Complete or partial protection from ZIKV replication were observed for six representative neutralizing mAbs. Following infusion with the indicated antibody or saline (Sham), ZIKV viral loads were measured in serum post challenge by RT-PCR daily until day 7. Viral load peaked at day 3 or 4. The six antibodies were tested in two sets of experiments, and a representative Sham group from one experiment is shown.
(C) Viral dissemination in brain, spleen, and lymphnodes (LNs) was assessed at day 3 post challenge for three of the most potent mAbs (black circles) as compared to the Sham group (red circles). Error bars indicate mean ± SEM.
See also STAR Methods.
DISCUSSION
The development of a ZIKV vaccine is a major public health priority. Since the geographic distributions of DENV and ZIKV overlap considerably, it is important to develop immunization strategies that elicit ZIKV-specific antibodies that do not cross-react with DENV and thus limit potential for ADE.54,55 Despite sequence and structural similarity between DENV and ZIKV, and some serological overlap between these viruses, ZIKV does maintain antigenic distinction from DENV. Previously, we determined that flavivirus-naive rhesus macaques did not demonstrate frequent or potent neutralization of DENV after ZIKV infection.34 From one of these animals, we sorted ZIKV-specific B cells using a sequential staining strategy with whole ZIKV. We were able to identify a set of strikingly potent ZIKV-specific neutralizing antibodies that targeted quaternary epitopes, with one mAb, rhMZ134-B, having an IC50 in the low ng/mL range. Analysis of the Ig-gene sequences indicated a low level of somatic hypermutation in these antibodies at 14 days post infection, in agreement with previous reports in humans.29,31
To delineate their specificities, we carried out competition experiments using previously identified mAbs known to target defined viral epitopes within FLE, EDE, DI/DII, and DIII. This revealed that our 11 neutralizing antibodies could be categorized into four antigenic groups, labeled A–D. Group A displayed a complicated antigenic competition profile competing with all control antibodies, and strong ZIKV-binding preference compared to monomeric E, indicating a quaternary epitope. Unfortunately, this group of antibodies eluded crystallization partly due to low affinity to E. Group B antibody epitopes overlapped with the known EDE specificity of EDE1-C8,42 and groups C and D mAbs defined two ZIKV inter-dimer epitopes that have not previously been described to our knowledge. The rhMZ100-C and rhMZ104-D antibody epitopes from groups C and D, respectively, bind two dimers in an almost equal manner. Similarly, mAb ZIKV-117 targets an epitope with some overlap with rhMZ100-C and rhMZ104-D inter-dimer epitopes. However, the ZIKV-117 epitope differs by primarily contacting one protomer.51
Our results demonstrate that antibody responses during acute ZIKV infection without prior DENV exposure target multiple protomers across the ZIKV dimer-dimer interface with a recognition pattern differing from that seen with DENV infection. Utilizing whole ZIKV in the sorting strategy may be a key factor contributing to these differences. We found the ZIKV specificity was due to a combination of the presence of the N67-glycan in DENV and sequence differences between DENV and ZIKV. Swapping the glycan loop (N154) of ZIKV with DENV has previously been reported to enhance the ZIKV neutralization by DII-FL antibodies,56 suggesting the significance of glycan in the antigenic structure of ZIKV. Here, using structural information, we found that the addition of the N67-glycan to ZIKV abrogated ZIKV neutralization. This glycan interference of an antigenic site is analogous to that seen in mAbs recognizing different influenza or HIV-1 strains where the presence of a glycan adversely affects antigenicity.49,57 Significantly, a study focused on characterizing potential ZIKV variants using sequential in vivo passage of a clinical isolate SZ-WIV01 (SW01) in neonatal mice identified a ZIKV variant harboring an E glycoprotein Asp-67-Asn mutation that resulted in about 100- to 1,000-fold increase in its neurovirulence.58
In this study, we observed genetically divergent antibodies that target overlapping epitopes, highlighting the fact that multiple pathways to generate these antibodies are possible, and equivalent mAbs likely can be generated in humans. Furthermore, the mAbs reported here have low SHM levels. The high potency of such mAbs is advantageous because of the relative ease of development and lack of genetic hurdles compared with antibodies with high SHM seen with influenza or HIV-1.59 Additionally, the inter-dimer epitopes can facilitate the design of ZIKV-specific immunogens with reduced cross-reactivity. This could improve the safety profile of vaccine candidates for DENV-endemic geographical regions where the potential for ADE is a major challenge.18 We postulate that neutralization by these groups of mAbs occurs via locking of the E proteins and thus a reduced ability of the E proteins to transition to a fusion-compatible ternary structure. The mAb cross-linking of E molecules likely blocks or inhibits the large conformational changes required for viral membrane fusion to facilitate flavivirus entry and will differ depending on the location of the cross-E epitope.
This study provides detailed epitope targeting of neutralizing antibodies specific to ZIKV that occurred 14 days after ZIKV infection in a flavivirus-naive scenario. Given the sophisticated nature of flavivirus inter-molecular contacts, there may be other epitopes elicited during ZIKV infection that have not yet been described, including those from antigenic group A, or located on pentameric or inter-raft vertices. Although potently neutralizing antibodies against ZIKV have been described previously, this work defines four ZIKV-specific neutralization antigenic sites, with two sites not previously described in ZIKV infection. Moreover, we have shown that divergent antibody pathways could be exploited during vaccine design to elicit these potent and specific antibodies. It will be of value to assess the presence of these antibody classes in DENV-experienced donors prior to or following ZIKV infection or vaccination to understand the potential of differential targeting of flaviviruses due to B cell priming. As the flavivirus immunology and vaccine development fields progress toward development of effective ZIKV vaccines, it is encouraging to see the potential for induction of either homologous or heterologous responses depending on patient history and immunogen selection. The mAb data specificity described here will be critical in the design of ZIKV vaccines and therapeutics that are effective, irrespective of the flavivirus exposure history.
Limitations of the study
Further studies to delineate the mechanism of mAb neutralization activity are needed. Structural definition of the group A mAbs would further inform the sites of vulnerability on ZIKV. Identification and characterization of equivalent human monoclonal antibodies would be highly informative. Further animal protection studies would inform the protective capacity of these mAbs, including neurocognitive protection,60 or blocking placental transmission in both naive or DENV-experienced backgrounds.61,62
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact M. Gordon Joyce (gjoyce@eidresearch.org).
Materials availability
All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and code availability
All data supporting the findings of this study are found within the paper and its Supplemental Information. The accession numbers for the Protein DataBank coordinates, and structure factors determined in this paper are PDB: 8EE8, 8EEE, 8EED, 8EE5, 8EEZ, 8EF1, 8EF0, 8EF2, and 8EF3, the GenBank accession numbers for the antibody sequences are GenBank: OR232903-OR232948.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT PARTICIPANT DETAILS
Rhesus macaque
Monoclonal antibodies were isolated from a flavivirus-naive macaque infected with Brazil ZK2015 (ZIKV-BR) previously described in McCracken et al., 2017.34 This study was approved by the Institutional Animal Care and Use Committee, and research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals. Experiments involving animals adhered to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition, National Research Council. The Walter Reed Army Institute of Research (WRAIR) is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Balb/C mice
Female Balb/c mice were purchased from commercial vendors and housed at Beth Israel Deaconess Medical Center. This study was approved by the Institutional Animal Care and Use Committee, and research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals.
In vivo protection studies
Female Balb/c mice were purchased from commercial vendors and housed at Beth Israel Deaconess Medical Center. The indicated macaque mAb was infused intravenously into groups of naive recipient Balb/c mice (N = 5/group) prior to ZIKV-BR challenge. Mice received 100 μL (200 μg) of a 2 mg/mL solution of purified mAb and 2 h after infusion, mice were challenged with 105 viral particles (VP) [102 plaque-forming units (PFU)] ZIKV-BR intravenously. RT-PCR assays were utilized to monitor viral loads.53 RNA was extracted from serum samples with a QIAcube HT (Qiagen). The wildtype ZIKV BeH815744 Cap gene was utilized as a standard. RNA was purified (Zymo Research). Log dilutions of the RNA standard were reverse transcribed and included with each RT-PCR assay. Viral loads were calculated as virus particles (VP) per mL with a sensitivity of 100 copies/ml.
Cell lines
D1–4G2–4-15 mouse hybridoma (ATCC #HB-112), C6/36 (ATCC #CRL-1660), Vero (ATCC #CCL-81), Expi293F (ThermoFisher Scientific #A14527), DS-2 (ThermoFisher Scientific #R69007), and U937-DC-SIGN (ATCC #CRL-3253, ATCC) cell lines were utilized in this study. These lines were verified to be authentic, using short tandem repeat profiling, morphology, and cytochrome c oxidase I testing, and free of contamination by mycoplasma prior to use.
Human samples
Plasma from DENV-experienced ZIKV-infected individuals were provided from ZIKABRA cohort. ZIKABRA is an observational cohort study in Brazil that recruited non-pregnant participants aged 18 years and above with a confirmed ZIKV infection and the study team investigators are provided below.69,70 All authors have complied with the ethical regulations regarding these studies. The study protocol and procedures have been reviewed and approved by WRAIR Institutional Review Board and Research Ethics Review Committee (WHO ERC), Protocol ID: ERC.0002786; Brazilian National Research Ethics Commission (CONEP)(CAAE: 62.518.016.6.1001.0008); Institutional Ethics and Research Committee of the Evandro Chagas National Institute of Infectious Diseases, Fiocruz, Rio de Janeiro (CAAE: 62.518.016.6.2002.5262); Institutional Ethics and Research Committee of the Aggeu Magalhães Research Center, Fiocruz, Recife (CAAE: 62.518.016.6.2001.5190) and Institutional Ethics and Research Committee of the Tropical Medicine Foundation, Manaus, Amazonas (CAAE: 62.518.016.6.2003.0005). Written informed consent was obtained from all participants. The investigators have adhered to the policies for protection of human participants as prescribed in AR 70–25. Healthy, flavivirus-naı¨ve plasmas originated from the WRAIR RV229 study and commercial source (Seracare). Other plasmas from ZIKV-infected donors were obtained commercially (AccuSet Zika performance panel, cat# 0845–0142, Seracare).
ZIKABRA Study Team (in alphabetical order): Abreu, André Luiz de (General Coordination of Public Health Laboratories (CGLAB/DAEVS/SVS/MS), Brasília-DF, Brazil); Bermudez, Ximena Pamela Diaz (Department of Public Health, University of Brasília, Brasília-DF, Brazil); Bôtto-Menezes, Camila Helena Aguiar (Department of Malaria, Tropical Medicine Foundation Doctor Heitor Vieira Dour-ado (FMT-HVD), Manaus, Amazonas, Brazil & School of Health Sciences, Amazonas State University (UEA), Manaus, Amazonas, Brazil); Brasil, Patrícia (Acute Febrile Illnesses Laboratory, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil); Brito, Carlos Alexandre Antunes (Clinical Hospital of Federal University, Department of Internal Medicine, Recife, Pernambuco, Brazil); Broutet, Nathalie Jeanne Nicole (Department of Sexual and Reproductive Health and Research, World Health Organization, Geneva, Switzerland; Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil); Castilho, Marcia da Costa (Department of Malaria, Tropical Medicine Foundation Doctor Heitor Vieira Dourado (FMT-HVD), Manaus, Amazonas, Brazil); Fernandes, Tatiana Jorge (Acute Febrile Illnesses Laboratory, Evandro Chagas National Institute of Infectious Diseases, Oswaldo Cruz Foundation, Rio de Janeiro, Rio de Janeiro, Brazil); Giozza, Silvana Pereira (Department of HIV/AIDS, Tuberculosis, Viral Hepatitis and Sexually Transmitted Infections, Health Surveillance Secretariat, Ministry of Health, Brazil); Habib, Ndema (Department of Sexual and Reproductive Health and Research, World Health Organization, Geneva, Switzerland); Kara, Edna Oliveira (Department of Sexual and Reproductive Health and Research, World Health Organization, Geneva, Switzerland); Lacerda, Marcus Vinicius Guimarães (Department of Malaria, Tropical Medicine Foundation Doctor Heitor Vieira Dourado (FMT-HVD), Manaus, Amazonas, Brazil & Instituto Leônidas & Maria Deane, Oswaldo Cruz Foundation, Manaus, Amazonas, Brazil); Lima, Morganna Costa (Department of Virology and Experimental Therapy, Institute Aggeu Magalhães, Oswaldo Cruz Foundation, Recife, Pernambuco, Brazil); Neto, Armando Menezes (Department of Virology and Experimental Therapy, Institute Aggeu Magalhães, Oswaldo Cruz Foundation, Recife, Pernambuco, Brazil); Pereira, Gerson Fernando (Department of HIV/AIDS, Tuberculosis, Viral Hepatitis and Sexually Transmitted Infections, Health Surveillance Secretariat, Ministry of Health, Brazil), Pimenta, Cristina (Department of HIV/AIDS, Tuberculosis, Viral Hepatitis and Sexually Transmitted Infections, Health Surveillance Secretariat, Ministry of Health, Brazil).
METHOD DETAILS
Preparation of ZIKV and DENV
C6/36 mosquito cells were grown in T75 flasks and infected with ZIKV strain (Paraiba_01 strain, GenBank KX280026) or DENV-2 (S16803, GenBank GU289914) at a multiplicity of infection of approximately 0.1 PFU/cell. The infected cell culture supernatant was harvested on day 5 post-infection. Cell debris was removed by centrifugation at 5,000 rpm for 30 min at 4°C. The supernatant was layered on top of a 30% sucrose solution containing 10 mM Tris, 100 mM NaCl, and 1 mM EDTA. The virus was pelleted by ultracentrifugation in a swinging-bucket rotor at 26,000 rpm for 4 h at 4°C to remove low-molecular-weight contaminants such as soluble proteins. The supernatant was removed, and the tubes were briefly left upside down on chromatography paper in order to remove excess liquid from the side of the tubes. The virus pellet was resuspended in phosphate-buffered saline. The purity of the viral preparations was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Sorting of ZIKV positive B cells
Approximately 10 million cryopreserved peripheral blood mononuclear cells (PBMCs) were obtained from a flavivirus-naïve, five-year-old male, rhesus macaque (#10U032) previously described.34 This animal was not previously infected with ZIKV, JEV, WNV, YFV, or DENV1–4 and this was confirmed by testing for antibodies by a sensitive screening virus neutralization assay prior to initial infection.34 The PBMC sample was collected 14 days following challenge with ZIKV (Brazil-ZKV2015 strain, GenBank KU497555). PBMCs were thawed in warm medium containing benzonase, then washed with phosphate-buffered saline (PBS) and stained for viability using Invitrogen Aqua Live/Dead stain. Cells were incubated at 4°C for 30 min with a cocktail of antibodies including CD3 BV510, CD14 BV510, and CD56 BV510 (BioLegend) as dump channel markers, and CD19 ECD (Beckman Coulter), CD20 APC-Cy7 (BioLegend), CD38 PE (NHP Reagent Resource, a gift from Diane Bolton) and CXCR5 PE-Cy7 (eBioscience) as positive gating markers. To obtain monoclonal antibodies that target quaternary epitopes, primary staining also included a 1/10 dilution of live whole ZIKV virions (Paraiba_01) produced in C6/36 cells (see above). ZIKV-reactive B cells were identified by secondary staining using 4G235 (Biovest) conjugated to APC (Thermofisher). Cells were selected by sorting based on negative expression of CD3, CD14 and CD56, positive expression of CD19, mid to high expression of CD38, and positive sequential staining with 4G2 (Figures 1A and 1B). Cells were sorted directly into lysis buffer (murine RNAse inhibitor (New England Biolabs), DTT and SuperScript III First Strand Buffer (ThermoFisher), Igepal (Sigma) and carrier RNA (Qiagen)) at one cell per well into polypropylene PCR plates using a FACSAria (Becton Dickinson) and stored at −80°C until subsequent reverse transcription.
Antibody sequencing and production
RNA from single B cells was reverse-transcribed using random primers and the SuperScriptIII kit (ThermoFisher). Antibody V (D) J genes were amplified from the cDNA by nested PCR, using the HotStar Taq DNA Polymerase kit (Qiagen) and a combination of primer sets.36–38 V (D) J gene assignment, somatic hypermutation and CDR3 determinations was performed with IgBlast.71 Antibody variable regions were synthesized and cloned (Genscript) into CMVR expression vectors (a gift from Kevin Saunders) between a murine Ig leader (GenBank DQ407610) and the constant regions of rhesus macaques IgG1 (GenBank AAF14058), Igκ (GenBank AAD02577) or Igλ (GenBank ADX62855).66 Variable regions for control antibodies EDE1-C8,42 Z3L1,45 2A10G6,64 and Z00443 were synthesized as mentioned above and cloned into CMVR expression vectors carrying human IgG1 (Swiss-Prot: P0DOX5.1), Igκ (Swiss-Prot: P01834.2) and Igλ (Swiss-Prot: P0DOY2.1) constant regions. Plasmids encoding heavy and light chains were co-transfected into Expi293F cells (ThermoFisher) according to the manufacturer’s instructions. After 5 days, antibodies were purified from cleared culture supernatants with Protein A agarose (ThermoFisher) using standard procedures, buffer exchanged to Phosphate-buffered saline (PBS) and quantified from A280 measurements. SDS-PAGE and Coomassie staining in both reducing and non-reducing conditions assessed purity and stability of the purified antibodies.
Fab production
Freshly prepared Fab digestion buffer containing 20 mM sodium phosphate, 10 mM EDTA and 20 mM of cysteine-HCl, pH 7.4 was added to papain slurry (Thermo Scientific) and incubated with rhMZ antibodies (IgGs) at 20 mg/mL. Reaction was allowed to proceed (for 5 h to overnight) in a shaker incubator at 37°C temperature and 100 rpm shaking speed. Resin was separated from the supernatant by centrifugation at 30003g. Digestion was assessed by SDS-PAGE, and upon completion, the reaction mixture was passed over protein A agarose (0.5–1 mL beads) 3 times and the final flow through was assessed by SDS-PAGE for purity.
Production of recombinant proteins
Recombinant ZIKV soluble E (sE) protein (amino acid 1–404) from strain PRVABC59 (GenBank KX087101) and DENV-2 E (amino acid 1–396) from strain 16681 (GenBank M84727) was produced with C-terminal AviTag and polyhistidine tags from Expi293F cells. The coding sequence for prM/sE was synthesized (Genscript) and cloned into the pcDNA3.4 vector (ThermoFisher) downstream of a murine Ig leader sequence. Following transient co-transfection with a human furin (GenBank BC012181) expression vector, mature E proteins were purified from cell culture supernatants using a Ni-NTA (Qiagen) affinity column. Cleavable twin-strep-tagged ZIKV E WT and L107C/A319C mutant (disulfide-stabilized dimer) version were also expressed from stably transfected S2 cells using the Drosophila Expression system (ThermoFisher) according to the manufacturer’s instructions. Briefly, a DNA fragment encoding for the first 405 residues of E from strain PRVABC59 was synthesized (Genscript) with a C-terminal human rhinovirus 3c protease (HRV-3c) cleavage site followed by a twin-strep tag (IBA) and cloned into the pMT-BiP vector (ThermoFisher). The L107C and A319C mutations were introduced using the Quikchange Lightning site-directed mutagenesis kit (Agilent). S2 cells were co-transfected with the pMT-BiP-ZIKV E WT and mutant expression vector and the pCoBlast selection vector at a 19:1 (w/w) ratio. Stably transfected cells were selected with Blasticidin and adapted to suspension and serum-free medium (Lonza Insect Xpress). ZIKV E expression was induced with 0.5mM CuSO4 and culture supernatants were harvested after 7 days. The insect-produced ZIKV E was purified on a StrepTactin XT column (IBA) following the manufacturer’s instructions followed by gel filtration on an Enrich SEC 650 column (Bio-Rad) or GE Sephadex S200 column to obtain pure monomeric (WT sE) or dimeric (mutant sE) ZIKV E proteins. C-terminally Avi-tagged versions of ZIKV (PRVABC59), DENV-1 (GenBank AAK60418), DENV-2 (GenBank AAC59274), DENV-3 (GenBank UCQ65256), DENV-4 (GenBank UXX63133), JEV (GenBank AAK11279) and YFV (GenBank AFQ32465) E proteins were also produced in the Drosophila S2 expression system, as described above, and purified by Ni-NTA affinity and size exclusion chromatography.
Biolayer interferometry mAb binding and competition assays
All real-time interactions between purified E proteins and antibodies were monitored on an Octet RED96 instrument (Pall ForteBio) at 30°C. Avi-tagged purified ZIKV or DENV-2 E proteins, biotinylated with the BirA biotinylation kit (Avidity), were diluted in kinetics buffer (0.1% [w/v] bovine serum albumin (BSA), 0.02% [v/v] Tween 20 in PBS; Pall ForteBio) and immobilized on streptavidin (SA) biosensors (Pall ForteBio) at ~50% of the sensor maximum binding capacity. Baseline was established in kinetics buffer. In the screening assay, loaded biosensors were then dipped into wells containing the antibodies diluted to 400 nM in kinetics buffer. Binding responses were measured after 450 s of association using the data analysis software 9.0 (Pall ForteBio). In the binding competition assay, sensors loaded with ZIKV E as described above were immersed into wells containing the first competing antibody at a concentration (ranging from 100 to 800 nM) necessary to reach binding saturation after 900 s. Next, biosensors were dipped into wells containing the second antibody, in presence of the first competing antibody, and binding was measured after 900 s of association. Residual binding signal of the second antibody was expressed as a percent of the signal obtained in presence of a non-competing control antibody (VRC01), assessed in parallel. As some competing antibodies did not reach saturation after the first 900 s association and continue to contribute to binding signal together with the second antibody, a set of controls were run independently with all first competing antibodies alone for 1800 s of association. The difference in signal obtained between t = 1800 s and t = 900 s was subtracted from the signal obtained in presence of the second antibody to generate a corrected residual binding signal. Antibodies were defined as competing when binding signal of the second antibody was reduced to less than 30% of its maximum binding capacity and non-competing when binding was greater than 70%. Intermediate competition was defined by binding levels of 30–70%. Control monoclonal antibodies included 2A10G6, expressed with a human Fc domain, Z3L1, EDE1-C8, and Z004, expressed and purified from Expi293F cells as described above. The HIV-1 specific VRC01 monoclonal antibody,63 also expressed in Expi293F cells, served as negative control.
Plasma competition assays were performed similarly to the mAb competition assays described above with the following modifications. Sensors loaded with ZIKV E were immersed into wells containing plasma from ZIKV infected macaques34 and humans, as well as control naive plasmas from the two species at dilutions (ranging from 1/10 to 1/200) necessary to reach near binding saturation after 900 s. Next, biosensors were dipped into wells containing the indicated monoclonal antibody, in presence of competing plasma, and binding was measured after 30 s of association. Residual binding signal of the monoclonal antibody was expressed as a percent of the signal obtained in presence of a non-competing matrix control of IgG-depleted human serum (BBI solutions), assessed in parallel. Binding of monoclonal antibodies was further corrected for the binding signal obtained with the matching plasma in the presence of negative control antibody (VRC01) assessed simultaneously. Finally, results were expressed as percentage of binding inhibition defined as the inverse of residual binding.
Measurement of antibody binding affinity
Determination of affinity constant was performed on the Octet RED96 instrument. Disulfide-stabilized ZIKV E was biotinylated at a 2:1 M ratio using EZ-link NHS-PEG4-biotin (ThermoFisher), following manufacturer’s instructions. A single buffer (1X kinetics buffer (Pall ForteBio)) was used for all dilution, baseline and dissociation steps. Streptavidin biosensors, loaded with ZIKV E dimer at ~50% of maximum binding capacity, were dipped into wells containing 2-fold serial dilutions of the antibody Fab fragments for 450 s with starting concentrations ranging from 1 to 10 μM. ZIKV sE:Fab complexes were then allowed to dissociate for 1200 s in buffer. After reference subtraction, binding kinetic constants were determined, from at least 4 concentrations of Fab, by fitting the curves to a 1:1 binding model using the data analysis software 9.0 (Pall ForteBio).
ELISA
Binding of antibodies to whole ZIKV or DENV-2 viruses described above was measured using a capture ELISA assay. ELISA plates were coated overnight at 4°C with the capture antibody 4G2 at 100 ng per well in borate saline pH 9.0 buffer. After washes in PBS-T (PBS with 0.05% [v/v] Tween 20), plates were blocked with 1% (v/v) normal goat serum, 0.25% (w/v) BSA, 0.1% (v/v) Tween 20 for 30 min at 37°C. Washes in PBS-T were performed after each subsequent step and all dilutions were made in blocking buffer. ZIKV and DENV were diluted and added at 50 μL per well and incubated for 2 h at 37°C. Serial 4-fold dilutions of antibodies starting at 20 μg/mL were added to the plate and incubated for 2 h at 37°C. Secondary HRP-conjugated antibodies anti-human and simian IgG were added for 1 h at 37°C and plates were developed using 3,3′,5,5′-Tetramethylbenzidine (TMB) peroxidase substrate (KPL) and read at 650 nm. The binding curves were fitted using a 4-parameter logistic regression model in the Prism 7 software (GraphPad).
Binding of antibodies to recombinant ZIKV or DENV-2 E proteins was also performed in a standard ELISA assay. ELISA plates were coated overnight at 4°C with 100 ng of purified ZIKV or DENV-2 E (as described above) in sodium bicarbonate/carbonate pH 9.4 buffer. Plates were then blocked with 5% (w/v) nonfat dry milk, 1% (w/v) BSA in PBS for 1 h at 37°C. Washes in between each step were performed with 0.1% (v/v) Triton X-100 in PBS. Serial 4-fold dilutions of antibodies starting at 20 μg/mL in 5% (v/v) Fetal Bovine Serum, 2% (w/v) BSA, 1% (v/v) Triton X-100 in PBS were added to the plate and incubated for 1 h at RT. Secondary HRP-conjugated antibodies, substrate and data analysis were as described above for the whole virus ELISA.
Multiplex antibody binding assay
A high-throughput bead-based antibody binding assay was performed72,73 with modifications to adapt to flavivirus antigens. Briefly, purified monoclonal antibodies were diluted and loaded into 384-well assay plates by use of a Biomek NXP automated liquid handler (Beckman Coulter). A cocktail of 7 flavivirus antigens and 1 control protein (HIV-1 antigen), produced internally, were covalently coupled to uniquely coded magnetic microspheres (Luminex) per manufacturer’s protocol and added to the plate in a final volume of 50 μL/well. Following a 2 h incubation with vigorous shaking, microspheres were washed using a magnetic 384-well automated plate washer (Bio-Tek) to remove unbound sample. Microspheres were then resuspended with 0.5 μg/mL mouse anti-human IgG-PE (Southern Biotech), vortexed for 1 min with a microplate vortex at 3,000 rpm, sonicated for 1 min and then incubated with vigorous shaking for 1 h. After a final wash to remove unbound detection reagent, microspheres were resuspended in 40 μL sheath fluid (Luminex). Data were collected on a Bio-Plex3D Suspension Array system (Bio-Rad) running xPONENT v.4.2 (Luminex). Signal to Noise (S/N) ratio were calculated by the dividing the MFI for each sample by an isotype control antibody (WRAIR-5002, a SARS-CoV-2 rhesus IgG1 antibody).
Shotgun mutagenesis epitope mapping
Epitope mapping was performed by shotgun mutagenesis.74 A ZIKV prM/E expression construct (strain ZIKV SPH2015) was subjected to high-throughput alanine scanning mutagenesis to generate a comprehensive library of individual mutations where each residue within prM/E was changed to alanine, with alanine mutated to serine. In total, 672 ZIKV prM/E mutants were generated (100% coverage), sequence confirmed, and arrayed into 384-well plates. Each prM/E mutant was transfected into HEK-293T cells and allowed to express for 22 h. Cells were fixed in 4% (v/v) paraformaldehyde (Electron Microscopy Sciences), permeabilized with 0.1% (w/v) saponin (Sigma-Aldrich) in PBS plus calcium and magnesium (PBS++), then incubated with purified mAbs diluted in PBS++, 10% (v/v) normal goat serum (NGS) (Sigma), 0.1% (v/v) saponin. Primary mAb screening concentrations were determined using an independent immunofluorescence titration curve against wild-type ZIKV prM/E to ensure that signals were within the linear range of detection. mAb binding was detected using 3.75μg/ml AlexaFluor488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) in 10% NGS/0.1% saponin. Cells were washed 3 times with PBS++/0.1% saponin followed by 2 washes in PBS. Mean cellular fluorescence was detected using a high throughput flow cytometer (HTFC, Intellicyt). mAb reactivities against each mutant prM/E clone were calculated relative to wild-type prM/E reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type prM/E-transfected controls. Mutations within clones were identified as critical to the mAb epitope if they did not support reactivity of the test mAb, but supported reactivity of other ZIKV mAbs. This counter-screen strategy facilitates the exclusion of prM/E mutants that are locally misfolded or have an expression defect.
Neutralization assays
Microneutralization (MN)
High-throughput ZIKV microneutralization (MN) assay was performed.53 Briefly, plasma or purified antibodies at 1 to 2 mg/mL were serially diluted 3-fold in 96-well micro-plates, and 100 μL of ZIKV containing 100 PFU were added to 100 μL of each serum dilution and incubated at 35°C for 2 h. Supernatants were then transferred to microtiter plates containing confluent Vero cell monolayers (World Health Organization, NICSC-011038011038). After incubation for 4 days, cells were fixed with absolute ethanol: methanol for 1 h at −20°C and washed three times with PBS. The pan flavivirus mAb 6B6-C1 conjugated to HRP (6B6-C1 was a gift from JT Roehrig, CDC) was then added to each well, incubated at 35°C for 2 h, and washed with PBS. Plates were washed, developed with TMB substrate for 50 min at room temperature, stopped with 1:25 phosphoric acid, and absorbance was read at 450 nm. Assays were validated using the following criteria: the average absorbance at 450 nm of three non-infected control wells had to be ≤0.5, and virus-only control wells had to be ≥0.9. Normalized absorbance values were calculated, and the concentration to achieve 50% neutralization (IC50) titer was calculated using an a five-parameter logistic regression analysis performed with the N-Parameter Logistic Regression (nplr) R package version 0.1–7.75 Reported IC50 values are geometric means calculated from at least two independent experiments.
FlowNT50
Serial dilutions of mAb or plasma were mixed with an equal volume of virus, diluted to achieve 10–15% infection of cells/well, and incubated for 1 h at 37°C. After 1 h of incubation, an equal volume of medium (RPMI-1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 1% L-glutamine (200 mM), and 1% non-essential amino acids (10 mM)) containing 5×104 U937-DC-SIGN cells were added to each serum-antibody mixture and incubated 18–20 h overnight in a 37°C, 5% CO2, humidified incubator. Following overnight incubation, the cells were fixed, permeabilized and immunostained with flavivirus group-reactive mouse mAb 4G2 (Envigo Bioproducts) and secondary polyclonal goat anti-mouse IgG PE-conjugated antibody. The percentage of infected cells was quantified on a BD Accuri C6 Plus flow cytometer (BD Biosciences). Reported IC50 values are the geometric means calculated from at least two independent experiments using a five-parameter logistic regression analysis, as described for the MN assay.
PRNT50
Serial dilutions of mAb were mixed with an equal volume of virus and incubated for 1 h at 37°C followed by infection of Vero-cell monolayers in triplicate. Plaques were visualized by staining with neutral red. Neutralization curves were graphed in Prism and reported IC50 values were calculated using a five-parameter logistic regression analysis, as described for the MN assay.
Reporter virus particle (RVP)
Neutralization of wildtype and mutant ZIKV (strain H/PF/2013) by mAbs was measured using a reporter virus particle (RVP) assay.76 Briefly, mAbs were serially diluted 5-fold from 50 μg and incubated with 100 μL of virus for 1 h at 37°C, after which 50 μL of target Vero cells (400,000 cells/ml) was added. Input virus dilution was calculated from titration experiments to ensure sufficient luciferase output within the linear portion of the titration curve. Cell only and virus only controls were included on each plate, and all mAbs (and virus-only) were run in triplicate. After a 48 h incubation, luciferase activity was measured, and neutralization curves were calculated by averaging luciferase units from triplicates, subtracting cell-only control background and calculating the percent difference in serum samples to virus-only controls. Data were fit by nonlinear regression using the asymmetric five-parameter logistic equation in GraphPad Prism. The 50%, 80% and 90% inhibitory dilutions (ID50, ID80 and ID90, respectively) were defined as the reciprocal sera dilution resulting in a 50%, 80% or 90% reduction in infectivity.
X-Ray crystallography and structure analysis
All proteins were crystallized by hanging-drop vapor diffusion at 273 K. Purified rhesus macaque Fabs and their complexes with ZIKV E glycoprotein were screened against a set of 1200 crystallization conditions using an Art Robbins Gryphon crystallization robot, and 0.2 μL drops. Crystal drops were observed daily using a Jan Scientific UVEX-PS with automated UV and bright field drop imaging. Initial crystallization conditions were optimized manually by mixing protein and reservoir solutions in 1:1 (v/v) ratios. Purified Fabs were concentrated to 7–10 mg/mL and used directly for crystallization screening. For complexes, Fabs and ZIKV E glycoprotein were mixed in an equimolar ratio at ~7 mg/mL and incubated at 4°C for 1 h prior to crystallization screening.
Crystals of the rhMZ103-A Fab were obtained at 8 mg/mL protein concentration and a reservoir solution containing 20% PEG 4000, 0.2M sodium acetate, 0.1M sodium citrate (pH 5.6). Crystals of the rhMZ107-B Fab were obtained at 7 mg/mL protein concentration and a reservoir solution containing 23.5% PEG 4000, 0.2 M (NH4)2SO4. Crystals of the rhMZ107-B Fab in complex with ZIKV E were obtained at 7 mg/mL protein concentration and a reservoir solution of 15% PEG 6000, 5% MPD, 0.1 M MES (pH 6.5). Crystals of the rhMZ100-C Fab were obtained at 8.1 mg/ml protein concentration and a reservoir solution containing 22.5% PEG 4000, 22.5% isopropanol, 0.1M sodium citrate (pH 5.6). Crystals of the rhMZ100-C Fab in complex with ZIKV E were obtained with a 7.5 mg/mL protein concentration and a reservoir solution of 12% PEG 8000, 0.2 M (NH4)2SO4, 0.1M Tris (pH 8.5). Crystals of the rhMZ104-D Fab were obtained at 8 mg/mL protein concentration and a reservoir solution containing 26% PEG 8000, 0.2M zinc acetate, 0.1M Tris-HCl (pH 8.5). Crystals of the rhMZ104-D Fab in complex with ZIKV E were obtained at 7 mg/mL protein concentration and a reservoir solution of 12% PEG 8000, 0.2 M (NH4)2SO4, 0.1M Tris-HCl (pH 8.5). Crystals of the rhMZ119-D Fab were obtained at 8.5 mg/mL protein concentration and a reservoir solution containing 18% PEG 8000, 0.2M calcium acetate hydrate, 0.1M sodium cacodylate trihydrate (pH 6.5). Crystals of the rhMZ119-D Fab in complex with ZIKV E were obtained at 6.0 mg/mL protein concentration and a reservoir solution containing 0.06M Nitrate Phosphate Sulfate, 0.1M Sodium HEPES and MOPS (acid) pH 7.5, 20% Ethylene glycol, 10% PEG 8000 + 2% w/v Benzamidine hydrochloride. All crystals were cryoprotected with mother liquor supplemented with 25% (v/v) glycerol prior to flash-cooling in liquid nitrogen. rhMZ119-D/ZIKV E complex formed soft and thin plate like crystals which did not survive high beam intensities and diffracted poorly (~10Å) at low beam intensity. Initial crystals of rhMZ107-B/ZIKV E complex also diffracted poorly with a limited resolution up to 8 Å. To improve the crystal quality crystallization condition, pH, ionic strength, and precipitant concentrations were varied. rhMZ119-D/ZIKV E complex was further screened for additives to improve the crystal quality. Addition of 2% w/v Benzamidine hydrochloride significantly improved the diffraction quality and produced thicker plate like crystals that were more resilient to the X-ray beam. The best crystals for rhMZ107-B/ZIKV E complex and rhMZ119-D/ZIKV E complex diffracted to a resolution of 3.5 and 3.58 Å, respectively.
Diffraction data were collected at Advanced Photon Source (APS), Argonne National Laboratory beamlines, and at National Synchrotron Light Source II (NSLS-II). Diffraction data for rhMZ103-A Fab, rhMZ100-C Fab, rhMZ100-C Fab/ZIKV E glycoprotein complex, rhMZ104-D Fab and rhMZ104-D Fab/ZIKV E glycoprotein complex were collected at APS 19-ID beamline to a final resolution of 1.87 Å, 2.19 Å, 2.8 Å, 2.48 Å and 2.8 Å, respectively, using a Pilatus 3 × 6M detector. Diffraction data for rhMZ107-B Fab crystals were collected at APS 24-ID-E beamline, and measured using a Dectris Eiger 16M PIXEL detector to a final resolution of 2.1 Å. Diffraction data for rhMZ107-B Fab/ZIKV E complex were collected at APS 22-BM beamline, and measured using a MX300HS CCD detector to a final resolution of 3.2 Å. Diffraction data for rhMZ119-D Fab were collected at APS 19-BM beamline, and measured using an ADSC Quantum 210r CCD detector to a final resolution of 1.67 Å. rhMZ119-D/ZIKV E complex diffraction data were collected at beamline NSLS-II AMX 17-ID-1 and measured using a Eiger 9M PIXEL detector to a final resolution of 3.58 Å. rhMZ119-D/ZIKV E complex diffraction data reduction and scaling were carried out utilizing the XDS based automated data processing pipeline, DIMPLE, at the AMX 17-ID-1 beamline. For all other crystals, diffraction data indexing, integration, and scaling were carried out using the HKL2000 suite.77 Data collection statistics are reported in Tables S4A and S4B.
All the crystal structures described in this study were solved by molecular replacement using PHASER, and iterative model building and refinement were performed in COOT68 and Phenix.78 Phenix xtriage was used to analyze all the scaled diffraction data output from HKL2000 and XDS. Primarily, data were analyzed for measurement value significance, completeness, asymmetric unit volume, and possible twinning and/or pseudotranslational pathologies. Fab datasets diffracted to resolution ranging from 1.67 Å to 2.48 Å, and phenix xtriage did not identify any data quality issue. PGT121-GL Fab structure (PDB code: 4FQQ) was used to solve the crystal structure of rhMZ103-A Fab. The rhMZ103-A Fab structure was used to solve the crystal structures of other Fabs reported in this study. Diffraction data for the rhMZ107-B/ZIKV E and rhMZ104-D complexes were mildly anisotropic whereas diffraction data for the rhMZ100-C/ZIKV E complex were significantly anisotropic and corrected using the UCLA Diffraction Anisotropy Server79 prior to structure solution. Although we processed the rhMZ100-C/ZIKV E complex data at 2.8 Å resolution, the resultant resolution after anisotropy correction was 2.9 Å. Phenix xtriage did not identify any other data quality issues for the rhMZ107-B/ZIKV E and rhMZ104-D/ZIKV E complexes. However, diffraction data for the rhMZ100-C complex crystal were significantly anisotropic and showed pseudo-merohedral twinning pathology at the 2-fold axis. To determine the structure of the Fab-ZIKV E structures, we divided the ZIKV E (PDB code: 5IRE) into smaller domains (DI, DII and DIII) and used a sequential search procedure. Crystal structures of the rhMZ Fabs determined in this study, were used as search models as is, or were further divided into Fv and Fc domains to find the molecular replacement solution. This search strategy was critical in finding the right solution for all the complexes. All the structures were refined using Phenix refine with positional, global isotropic B-factor refinement and defined TLS groups. Manual model building was performed in COOT. The final stages of refinement were performed with release of all non-crystallographic symmetry (NCS) restraints. The Ramachandran plot as determined by MOLPROBITY80 showed 90–97% of all residues in favored regions and 98–100% of all residues in the allowed regions. Data collection and refinement statistics are reported in Tables S4A and S4B. We also compared our structures, using Rfree as the basis, to the other structures of similar resolution in the RCSB database and our structures were better than 14.5 to 88.5% of the reported structures. The percentile values from this analysis for the individual structures are also reported in the Tables S4A and S4B. Interactive surfaces were analyzed using PISA (www.ebi.ac.uk/pdbe/pisa/). Structure figures were prepared using PyMOL (The PyMOL Molecular Graphics System (DeLano Scientific)).
Modeling differences between surface sites of dengue and ZIKV E protomers
An initial multiple sequence alignment including a recent Brazil ZIKV E (GenBank accession: AMA12087) and E of DENV1–4 (GenBank accession: DENV1, NP_059433; DENV2, NP_056776; DENV3, YP_001621843; DENV4, NP_073286) was established using MAFFT.81 After manual adjustment of the alignment, differences between amino acids on corresponding sites of E of ZIKV and DENV1–4 were calculated as BOLOSUM62 score82 and mapped on ZIKV E (PDB: 5IRE) for illustration. A residue pair is considered as similar (or identical) if the BOLOSUM62 score is greater than or equal to 1; otherwise the residue pair was considered different.
QUANTIFICATION AND STATISTICAL ANALYSIS
Binding experiments are presented as the mean values ±s.e.m. calculated from two independent experiments. Neutralization is the geometric mean of the IC50 values calculated using five-parameter regression, performed in R (version 3.5.1), from at least two independent experiments performed in triplicate. Independent groups were compared using Mann-Whitney tests, and matched paired data from wildtype (WT) and D67N-A69T ZIKV mutant RVP assays were analyzed by Wilcoxon signed rank test. Spearman’s coefficient was used for correlation analyses. The threshold for statistical significance was set to p < 0.05 for all analyses. Data were graphed using PRISM software (version 7, Graphpad Software, La Jolla, CA, USA).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-Zika virus E antibody, rhMZ100-C | This manuscript | OR232903 & OR232904 |
| Anti-Zika virus E antibody, rhMZ101-B | This manuscript | OR232905 & OR232906 |
| Anti-Zika virus E antibody, rhMZ103-A | This manuscript | OR232907 & OR232908 |
| Anti-Zika virus E antibody, rhMZ104-D | This manuscript | OR232909 & OR232910 |
| Anti-Zika virus E antibody, rhMZ105 | This manuscript | OR232911 & OR232912 |
| Anti-Zika virus E antibody, rhMZ106 | This manuscript | OR232913 & OR232914 |
| Anti-Zika virus E antibody, rhMZ107-B | This manuscript | OR232915 & OR232916 |
| Anti-Zika virus E antibody, rhMZ113 | This manuscript | OR232917 & OR232918 |
| Anti-Zika virus E antibody, rhMZ115 | This manuscript | OR232919 & OR232920 |
| Anti-Zika virus E antibody, rhMZ118 | This manuscript | OR232921 & OR232922 |
| Anti-Zika virus E antibody, rhMZ119-D | This manuscript | OR232923 & OR232924 |
| Anti-Zika virus E antibody, rhMZ120 | This manuscript | OR232925 & OR232926 |
| Anti-Zika virus E antibody, rhMZ121-A | This manuscript | OR232927 & OR232928 |
| Anti-Zika virus E antibody, rhMZ123-A | This manuscript | OR232929 & OR232930 |
| Anti-Zika virus E antibody, rhMZ124-D | This manuscript | OR232931 & OR232932 |
| Anti-Zika virus E antibody, rhMZ125 | This manuscript | OR232933 & OR232934 |
| Anti-Zika virus E antibody, rhMZ129 | This manuscript | OR232935 & OR232936 |
| Anti-Zika virus E antibody, rhMZ130 | This manuscript | OR232937 & OR232938 |
| Anti-Zika virus E antibody, rhMZ132 | This manuscript | OR232939 & OR232940 |
| Anti-Zika virus E antibody, rhMZ133-C | This manuscript | OR232941 & OR232942 |
| Anti-Zika virus E antibody, rhMZ134-B | This manuscript | OR232943 & OR232944 |
| Anti-Zika virus E antibody, rhMZ136 | This manuscript | OR232945 & OR232946 |
| Anti-Zika virus E antibody, rhMZ140 | This manuscript | OR232947 & OR232948 |
| Human recombinant EDE1-C8 | Barba-Spaeth et al., 2017 | Barba-Spaeth et al., 201742 |
| Human recombinant Z004 | Robbiani et al., 2017 | Robbiani et al., 201743 |
| Human recombinant Z3L1 | Wang et al., 2016 | Wang et al., 201645 |
| Human recombinant VRC01 | Wu et al., 2010 | Wu et al., 201063 |
| Mouse recombinant 2A10G6/humanized Fc | Deng et al., 2011/Dussupt et al., 2020 | Deng et al., 201164 Dussupt et al., 202024 |
| Mouse anti-pan-flavivirus E 6B6-C1 | J.T. Roehrig, CDC | N/A |
| Mouse anti-pan-flavivirus E D1-4G2-4-15 (4G2) | Biovest | N/A |
| Mouse anti-monkey IgG, HRP conjugated | Southern Biotech | Cat# 4700-05 |
| Mouse anti-human IgG, HRP conjugated | Southern Biotech | Cat# 9040-05 |
| Mouse anti-human CD3, BV510 conjugated | BD Biosciences | Cat# 740187 |
| Mouse anti-human CD14, BV510 conjugated | Biolegend | Cat# 301841; RRID:AB_2561379 |
| Mouse anti-human CD56, BV510 conjugated | Biolegend | Cat# 318339; RRID:AB_2561385 |
| Mouse anti-human CD19, ECD conjugated | Beckman Coulter | Cat# IM2708U; RRID:AB_130854 |
| Mouse anti-human CD20, APC-Cy7 conjugated | Biolegend | Cat# 302313; RRID:AB_314261 |
| Mouse anti-human CD38, PE conjugated | NHP Reagent Resource,gift from Dr. Diane Bolton | N/A |
| Mouse anti-human CD185, PE-Cy7 conjugated | eBioscience | Cat# 25-9185-42; RRID:AB_2573540 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Stbl3 competent cells | ThermoFisher Scientific | Cat# C737303 |
| Top10 competent cells | ThermoFisher Scientific | Cat# C404010 |
| E.coli DH5 alpha | ThermoFisher Scientific | Cat# 18265017 |
| ZIKV Paraiba_01 | S. Whitehead, NIAID | GenBank# KX280026 |
| ZIKV PRVABC59 | CDC | GenBank# KX087101 |
| ZIKV Thailand/2014 | WRAIR/AFRIMS | GenBank# KM851039 |
| ZIKV Uganda/1947 | Kind gift from S. Whitehead, NIAID | GenBank# NC_012532 |
| ZIKV-BR; Brazil/ZIKV2015 | Cugola et al.65 | GenBank# KU497555 Cugola etal.,201665 |
| ZIKV BeH815744 | Larocca et al.53 | GenBank# KU365780 Larocca et al. 201653 |
| DENV-2 S16803 | Kind gift from WRAIR/AFRIMS | GenBank# GU289914 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Benzonase nuclease | Novagen | Cat# 70664-3 |
| Aqua Live/Dead stain | ThermoFisher Scientific | Cat# L34957 |
| Igepal | Sigma-Aldrich | Cat# I8896 |
| Murine RNAse inhibitor | New England Biolabs | Cat# M0314L |
| Carrier RNA | ThermoFisher Scientific | Cat# 4382878 |
| Protein A agarose | ThermoFisher Scientific | Cat# 20334 |
| NiNTA agarose | Qiagen | Cat# 30210 |
| Streptactin XT superflow | IBA | Cat# 2-4010-025 |
| 1-Step Ultra TMB-ELISA Substrate | ThermoFisher Scientific | Cat# 34029 |
| Bovine serum albumin | Sigma-Aldrich | Cat# A8327 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Alexa Fluor 647 Antibody Labeling Kit | ThermoFisher Scientific | Cat# A20186 |
| SuperScriptIII kit | ThermoFisher Scientific | Cat# 18080085 |
| HotStar Taq Plus Polymerase kit | Qiagen | Cat# 203207 |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | Cat# E0554S |
| Quickchange Site-Directed Mutagenesis kit | Agilent | Cat# 210518 |
| Expi293 Expression system | ThermoFisher Scientific | Cat# A14635 |
| Drosophila Expression System | ThermoFisher Scientific | Cat# K513001 |
| BirA biotinylation kit | Avidity | Cat# bulk BirA |
| EZ-Link NHS-PEG4 Biotinylation Kit | ThermoFisher Scientific | Cat# 21455 |
| LAL Endotoxin detection kit | Lonza | Cat# 50-647U |
| QIAcube HT | Qiagen | Cat# 9001793 |
| Cador Pathogen 96 QIAcube HT Kit | Qiagen | Cat# 54161 |
|
| ||
| Deposited data | ||
|
| ||
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ100-C in complex with ZIKV E glycoprotein | This manuscript | PDB: 8EE8 |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ104-D in complex with ZIKV E glycoprotein | This manuscript | PDB: 8EEE |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ107-B in complex with ZIKV E glycoprotein | This manuscript | PDB: 8EED |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ119-D in complex with ZIKV E glycoprotein | This manuscript | PDB: 8EE5 |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ100-C | This manuscript | PDB: 8EEZ |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ103-A | This manuscript | PDB: 8EF1 |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ104-D | This manuscript | PDB: 8EF0 |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ107-B | This manuscript | PDB: 8EF2 |
| Crystal structure of an NHP anti-ZIKV neutralizing antibody rhMZ119-D | This manuscript | PDB: 8EF3 |
| Anti-Zika virus E antibody, rhMZ100-C Heavy chain | This manuscript | GenBank: OR232903 |
| Anti-Zika virus E antibody, rhMZ100-C | This manuscript | GenBank: OR232904 |
| Anti-Zika virus E antibody, rhMZ101-B Heavy chain | This manuscript | GenBank: OR232905 |
| Anti-Zika virus E antibody, rhMZ101-B | This manuscript | GenBank: OR232906 |
| Anti-Zika virus E antibody, rhMZ103-A Heavy chain | This manuscript | GenBank: OR232907 |
| Anti-Zika virus E antibody, rhMZ103-A | This manuscript | GenBank: OR232908 |
| Anti-Zika virus E antibody, rhMZ104-D Heavy chain | This manuscript | GenBank: OR232909 |
| Anti-Zika virus E antibody, rhMZ104-D | This manuscript | GenBank: OR232910 |
| Anti-Zika virus E antibody, rhMZ105 Heavy chain | This manuscript | GenBank: OR232911 |
| Anti-Zika virus E antibody, rhMZ105 | This manuscript | GenBank: OR232912 |
| Anti-Zika virus E antibody, rhMZ106 Heavy chain | This manuscript | GenBank: OR232913 |
| Anti-Zika virus E antibody, rhMZ106 | This manuscript | GenBank: OR232914 |
| Anti-Zika virus E antibody, rhMZ107-B Heavy chain | This manuscript | GenBank: OR232915 |
| Anti-Zika virus E antibody, rhMZ107-B | This manuscript | GenBank: OR232916 |
| Anti-Zika virus E antibody, rhMZ113 Heavy chain | This manuscript | GenBank: OR232917 |
| Anti-Zika virus E antibody, rhMZ113 | This manuscript | GenBank: OR232918 |
| Anti-Zika virus E antibody, rhMZ115 Heavy chain | This manuscript | GenBank: OR232919 |
| Anti-Zika virus E antibody, rhMZ115 | This manuscript | GenBank: OR232920 |
| Anti-Zika virus E antibody, rhMZ118 Heavy chain | This manuscript | GenBank: OR232921 |
| Anti-Zika virus E antibody, rhMZ118 | This manuscript | GenBank: OR232922 |
| Anti-Zika virus E antibody, rhMZ119-D Heavy chain | This manuscript | GenBank: OR232923 |
| Anti-Zika virus E antibody, rhMZ119-D | This manuscript | GenBank: OR232924 |
| Anti-Zika virus E antibody, rhMZ120 Heavy chain | This manuscript | GenBank: OR232925 |
| Anti-Zika virus E antibody, rhMZ120 | This manuscript | GenBank: OR232926 |
| Anti-Zika virus E antibody, rhMZ121-A Heavy chain | This manuscript | GenBank: OR232927 |
| Anti-Zika virus E antibody, rhMZ121-A | This manuscript | GenBank: OR232929 |
| Anti-Zika virus E antibody, rhMZ123-A Heavy chain | This manuscript | GenBank: OR232930 |
| Anti-Zika virus E antibody, rhMZ123-A | This manuscript | GenBank: OR232931 |
| Anti-Zika virus E antibody, rhMZ124-D Heavy chain | This manuscript | GenBank: OR232932 |
| Anti-Zika virus E antibody, rhMZ124-D | This manuscript | GenBank: OR232933 |
| Anti-Zika virus E antibody, rhMZ125 Heavy chain | This manuscript | GenBank: OR232934 |
| Anti-Zika virus E antibody, rhMZ125 | This manuscript | GenBank: OR232935 |
| Anti-Zika virus E antibody, rhMZ129 Heavy chain | This manuscript | GenBank: OR232936 |
| Anti-Zika virus E antibody, rhMZ129 | This manuscript | GenBank: OR232937 |
| Anti-Zika virus E antibody, rhMZ130 Heavy chain | This manuscript | GenBank: OR232938 |
| Anti-Zika virus E antibody, rhMZ130 | This manuscript | GenBank: OR232939 |
| Anti-Zika virus E antibody, rhMZ132 Heavy chain | This manuscript | GenBank: OR232940 |
| Anti-Zika virus E antibody, rhMZ132 | This manuscript | GenBank: OR232941 |
| Anti-Zika virus E antibody, rhMZ133-C Heavy chain | This manuscript | GenBank: OR232942 |
| Anti-Zika virus E antibody, rhMZ133-C | This manuscript | GenBank: OR232943 |
| Anti-Zika virus E antibody, rhMZ134-B Heavy chain | This manuscript | GenBank: OR232944 |
| Anti-Zika virus E antibody, rhMZ134-B | This manuscript | GenBank: OR232945 |
| Anti-Zika virus E antibody, rhMZ136 Heavy chain | This manuscript | GenBank: OR232946 |
| Anti-Zika virus E antibody, rhMZ136 | This manuscript | GenBank: OR232947 |
| Anti-Zika virus E antibody, rhMZ140 Heavy chain | This manuscript | GenBank: OR232948 |
| Anti-Zika virus E antibody, rhMZ140 | This manuscript | GenBank: OR232949 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| D1-4G2-4-15 mouse hybridoma | ATCC | Cat# HB-112; RRID:CVCL_J890 |
| C6/36 | ATCC | Cat# CRL-1660; RRID:CVCL_Z230 |
| Vero | ATCC | Cat# CCL-81; RRID:CVCL_0059 |
| Expi293F | ThermoFisher Scientific | Cat# A14527 |
| S2 | ThermoFisher Scientific | Cat# R69007 |
| U937-DC-SIGN | ATCC | Cat# CRL-3253; RRID:CVCL_2Z95 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Balb/c mice | Jackson Laboratories | Cat# 000651; RRID:IMSR_JAX:000651 |
|
| ||
| Recombinant DNA | ||
|
| ||
| Macaque IgG1, Igkand Igl expression vectors | Saunders et al.66 | Saunders et al., 201566 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for antibody nested PCR | Sundling et al.36 | Sundling et al., 201236 |
| Primers for antibody nested PCR | Meng et al.37 | Meng et al., 201537 |
| Primers for antibody nested PCR | Mason et al.38 | Mason et al., 201638 |
| ZIKV.Cap.RT.probe AGTTCAAG AAAGATCTGGCTG | Larocca et al.53 | Larocca et al. 201653 |
| ZIKV.Cap.RT.fwd GGAAAAAAGA GGCTATGGAAATAATAAAG | Larocca et al.53 | Larocca et al. 201653 |
| ZIKV.Cap.RT.rev CTCCTTCCTA GCATTGATTATTCTCA | Larocca et al.53 | Larocca et al. 201653 |
|
| ||
| Software and algorithms | ||
|
| ||
| Biorender | Biorender | https://biorender.com/ |
| IgBlast | Ye et al.71 | https://www.ncbi.nlm.nih.gov/igblast/ Ye et al.71 |
| Protein Repair One-Stop Shop (PROSS) server | Goldenzweig et al., 201667 | https://pross.weizmann.ac.il/step/pross-terms/ Goldenzweig et al., 201667 |
| Octet Data Analysis software | ForteBio | v11.1 |
| GraphPad Prism | Graphpad | v9.0 https://www.graphpad.com/ |
| PyMol | Schrodinger | v2.3.2 |
| COOT | Emsley and Cowtan.68 | http://bernhardcl.github.io/coot/ Emsley and Cowtan, 200468 |
| SnapGene | Insightful Science | https://www.snapgene.com/ |
|
| ||
| Other | ||
|
| ||
| Superdex 200 increase 10/300 GL | Cytiva | Cat# 28990944 |
| Enrich SEC 650 column | Bio-Rad | Cat# 7801650 |
Using whole ZIKV as a probe, we identify potent ZIKV-specific neutralizing antibodies
Neutralizing mAbs target four distinct epitopes at the E dimer-dimer interface
The four ZIKV-specific epitopes are prevalent in ZIKV infection in humans
mAbs from all four antigenic groups provide protection against ZIKV infection
ACKNOWLEDGMENTS
We thank Kevin Saunders for the gift of the rhesus mAb expression vectors, Matthew Creegan and Michael Eller for help with FACS sorting, and Caitlin Kuklis and Qiong Chen for technical support. X-ray data were collected at three beamlines at the Advanced Photon Source, Argonne National Laboratory, including Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences, under contract no. W-31–109-Eng-38. The Structural Biology Center (SBC) and Northeastern Collaborative Access Team (NE-CAT) beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403) at the Advanced Photon Source, Argonne National Laboratory. SBC-CAT is operated by UChicago Argonne, LLC, for the DOE, Office of Biological and Environmental Research under contract DE-AC02–06CH11357. This research also used the AMX 17-ID-1 beamline of the National Synchrotron Light Source II, a DOE Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract no. DE-SC0012704. This work was funded by the US Department of Defense, the Defense Health Agency, and the Department of the Army and supported through a cooperative agreement (W81XWH-07–2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. and the US Department of Defense, as well as through the Military Infectious Disease Research Program. In addition, funding was supported by NIH contract HHSN75N93019C00073 to B.J.D. and R01AI155983 to S.J.K. and K.M. This material has been reviewed by the Walter Reed Army Institute of Research. Research was conducted under an approved animal use protocol in an AAALACi-accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition. There is no objection to its presentation and/or publication. The authors thank the ZIKABRA study team, whose details are listed in the supplemental information and at https://eidresearch.org/ZIKABRAStudyTeam. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense.
Footnotes
DECLARATION OF INTERESTS
S.J.K., V.D., G.D., K.M., N.M., D.H.B., M.G.J., R.S.S., and R.G.J. are named inventors on a PCT patent application WO 2019/209974 describing ZIKV neutralizing antibodies and their use. D.H.B. has received grants from Novavax and personal fees from IGM Biosciences. M.H., A.A., E.D., and B.J.D. are employees of Integral Molecular. B.J.D. is also a shareholder of the company.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112942.
REFERENCES
- 1.Lazear HM, and Diamond MS (2016). Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 90, 4864–4875. 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao-Lormeau VM, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. (2016). Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539. 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carteaux G, Maquart M, Bedet A, Contou D, Brugières P, Fourati S, Cleret de Langavant L, de Broucker T, Brun-Buisson C, Leparc-Goffart I, and Mekontso Dessap A (2016). Zika Virus Associated with Meningoencephalitis. N. Engl. J. Med. 374, 1595–1596. 10.1056/NEJMc1602964. [DOI] [PubMed] [Google Scholar]
- 4.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, et al. (2016). Zika Virus Associated with Microcephaly. N. Engl. J. Med. 374, 951–958. 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 5.Macnamara FN (1954). Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans. R. Soc. Trop. Med. Hyg. 48, 139–145. [DOI] [PubMed] [Google Scholar]
- 6.Dick GW, Kitchen SF, and Haddow AJ (1952). Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 46, 509–520. [DOI] [PubMed] [Google Scholar]
- 7.Ayres CFJ (2016). Identification of Zika virus vectors and implications for control. Lancet Infect. Dis. 16, 278–279. 10.1016/S1473-3099(16)00073-6. [DOI] [PubMed] [Google Scholar]
- 8.Mead PS, Duggal NK, Hook SA, Delorey M, Fischer M, Olzenak McGuire D, Becksted H, Max RJ, Anishchenko M, Schwartz AM, et al. (2018). Zika Virus Shedding in Semen of Symptomatic Infected Men. N. Engl. J. Med. 378, 1377–1385. 10.1056/NEJMoa1711038. [DOI] [PubMed] [Google Scholar]
- 9.Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonc a, M.C.L., de Oliveira L, et al. (2016). Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 16, 653–660. 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 10.Musso D, Roche C, Robin E, Nhan T, Teissier A, and Cao-Lormeau VM (2015). Potential sexual transmission of Zika virus. Emerg. Infect. Dis. 21, 359–361. 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasquez AM, Sapiano MRP, Basavaraju SV, Kuehnert MJ, and Rivera-Garcia B (2016). Survey of Blood Collection Centers and Implementation of Guidance for Prevention of Transfusion-Transmitted Zika Virus Infection–Puerto Rico. MMWR Morb. Mortal. Wkly. Rep. 65, 375–378. 10.15585/mmwr.mm6514e1. [DOI] [PubMed] [Google Scholar]
- 12.Wikan N, and Smith DR (2016). Zika virus: history of a newly emerging arbovirus. Lancet Infect. Dis. 16, e119–e126. 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 13.Heukelbach J, Alencar CH, Kelvin AA, de Oliveira WK, and Pamplona de Góes Cavalcanti, L. (2016). Zika virus outbreak in Brazil. J. Infect. Dev. Ctries. 10, 116–120. 10.3855/jidc.8217. [DOI] [PubMed] [Google Scholar]
- 14.Gullo G, Scaglione M, Cucinella G, Riva A, Coldebella D, Cavaliere AF, Signore F, Buzzaccarini G, Spagnol G, Laganà AS, et al. (2022). Congenital Zika Syndrome: Genetic Avenues for Diagnosis and Therapy, Possible Management and Long-Term Outcomes. J. Clin. Med. 11, 1351. 10.3390/jcm11051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong P, Hennessey M, Adams M, Cherry C, Chiu S, Harrist A, Kwit N, Lewis L, McGuire DO, Oduyebo T, et al. (2016). Travel-Associated Zika Virus Disease Cases Among U.S. Residents–United States, January 2015-February 2016. MMWR Morb. Mortal. Wkly. Rep 65, 286–289. 10.15585/mmwr.mm6511e1. [DOI] [PubMed] [Google Scholar]
- 16.Graham SD, Tu HA, McElvany BD, Graham NR, Grinyo A, Davidson E, Doranz BJ, Diehl SA, de Silva AM, and Markmann AJ (2021). A Novel Antigenic Site Spanning Domains I and III of the Zika Virus Envelope Glycoprotein Is the Target of Strongly Neutralizing Human Monoclonal Antibodies. J. Virol. 95, e02423–20. 10.1128/JVI.02423-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, et al. (2016). Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. USA 113, 7852–7857. 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, and Harris E (2017). Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932. 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramıŕez JO, Carrasquilla G, et al. (2015). Efficacy of a tetravalent dengue vaccine in children in Latin America. N. Engl. J. Med. 372, 113–123. 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- 20.Hadinegoro SR, Arredondo-García JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HIH, Reynales H, Limkittikul K, Rivera-Medina DM, et al. (2015). Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N. Engl. J. Med. 373, 1195–1206. 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 21.Burton DR, and Hangartner L (2016). Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol. 34, 635–659. 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barouch DH, Thomas SJ, and Michael NL (2017). Prospects for a Zika Virus Vaccine. Immunity 46, 176–182. 10.1016/j.immuni.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Wang R, Gao F, Li M, Liu J, Wang J, Hong W, Zhao L, Wen Y, Yin C, et al. (2017). Delineating antibody recognition against Zika virus during natural infection. JCI Insight 2, e93042. 10.1172/jci.insight.93042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dussupt V, Sankhala RS, Gromowski GD, Donofrio G, De La Barrera RA, Larocca RA, Zaky W, Mendez-Rivera L, Choe M, Davidson E, et al. (2020). Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor. Nat. Med. 26, 228–235. 10.1038/s41591-019-0746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierson TC, and Diamond MS (2012). Degrees of maturity: the complex structure and biology of flaviviruses. Curr. Opin. Virol. 2, 168–175. 10.1016/j.coviro.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey FA, Stiasny K, Vaney MC, Dellarole M, and Heinz FX (2018). The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 19, 206–224. 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, and Kuhn RJ (2016). The 3.8 A resolution cryo-EM structure of Zika virus. Science 352, 467–470. 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Li S, Du L, Wang C, Zou P, Hong B, Yuan M, Ren X, Tai W, Kong Y, et al. (2017). Neutralization of Zika virus by germline-like human monoclonal antibodies targeting cryptic epitopes on envelope domain III. Emerg. Microb. Infect. 6, e89. 10.1038/emi.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826. 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Yan J, and Gao GF (2017). Monoclonal Antibodies against Zika Virus: Therapeutics and Their Implications for Vaccine Design. J. Virol. 91, e01049–17. 10.1128/JVI.01049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers TF, Goodwin EC, Briney B, Sok D, Beutler N, Strubel A, Nedellec R, Le K, Brown ME, Burton DR, and Walker LM (2017). Zika virus activates de novo and cross-reactive memory B cell responses in dengue-experienced donors. Sci. Immunol. 2, eaan6809. 10.1126/sciimmunol.aan6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvert AE, Horiuchi K, Boroughs KL, Ong YT, Anderson KM, Biggerstaff BJ, Stone M, Simmons G, Busch MP, and Huang CYH (2021). The Specificity of the Persistent IgM Neutralizing Antibody Response in Zika Virus Infections among Individuals with Prior Dengue Virus Exposure. J. Clin. Microbiol. 59, e0040021. 10.1128/JCM.00400-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins MH, McGowan E, Jadi R, Young E, Lopez CA, Baric RS, Lazear HM, and de Silva AM (2017). Lack of Durable Cross-Neutralizing Antibodies Against Zika Virus from Dengue Virus Infection. Emerg. Infect. Dis. 23, 773–781. 10.3201/eid2305.161630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCracken MK, Gromowski GD, Friberg HL, Lin X, Abbink P, DeLa Barrera R, Eckles KH, Garver LS, Boyd M, Jetton D, et al. (2017). Impact of prior flavivirus immunity on Zika virus infection in rhesus macaques. PLoS Pathog. 13, e1006487. 10.1371/journal.ppat.1006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henchal EA, Gentry MK, McCown JM, and Brandt WE (1982). Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31, 830–836. [DOI] [PubMed] [Google Scholar]
- 36.Sundling C, Phad G, Douagi I, Navis M, and Karlsson Hedestam GB(2012). Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J. Immunol. Methods 386, 85–93. 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Meng W, Li L, Xiong W, Fan X, Deng H, Bett AJ, Chen Z, Tang A, Cox KS, Joyce JG, et al. (2015). Efficient generation of monoclonal antibodies from single rhesus macaque antibody secreting cells. mAbs 7, 707–718. 10.1080/19420862.2015.1051440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O’Dell S, Lusvarghi S, Bewley CA, et al. (2016). Targeted Isolation of Antibodies Directed against Major Sites of SIV Env Vulnerability. PLoS Pathog. 12, e1005537. 10.1371/journal.ppat.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, et al. (2018). Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation. Immunity 48, 339–349.e5. 10.1016/j.immuni.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corcoran MM, Phad GE, Vázquez Bernat N, Stahl-Hennig C, Sumida N, Persson MAA, Martin M, and Karlsson Hedestam GB (2016). Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 7, 13642. 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. (2015). A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177. 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I,Sharma A, Simon-Lorière E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. (2016). Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53. 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- 43.Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, Schaefer-Babajew D, Avila-Rios S, Nogueira L, Patel R, et al. (2017). Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 169, 597–609.e11. 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, et al. (2015). Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113. 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, Wong G, Peng R, Liu S, Li J, et al. (2016). Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci. Transl. Med. 8, 369ra179. 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Kostyuchenko VA, Ng TS, Lim XN, Ooi JSG, Lambert S, Tan TY, Widman DG, Shi J, Baric RS, and Lok SM (2016). Neutralization mechanism of a highly potent antibody against Zika virus. Nat. Commun. 7, 13679. 10.1038/ncomms13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412. 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. (2016). Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623. 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. (2016). Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540, 443–447. 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, et al. (2017). A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun. 8, 14722. 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, and Rossmann MG (2006). Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124, 485–493. 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 53.Larocca RA, Abbink P, Peron JPS, Zanotto P.M.d.A., Iampietro MJ, Badamchi-Zadeh A, Boyd M, Ng’ang’a D, Kirilova M, Nityanandam R, et al. (2016). Vaccine protection against Zika virus from Brazil. Nature 536, 474–478. 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. (2016). Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 17, 1102–1108. 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, et al. (2017). Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356, 175–180. 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goo L, DeMaso CR, Pelc RS, Ledgerwood JE, Graham BS, Kuhn RJ, and Pierson TC (2018). The Zika virus envelope protein glycan loop regulates virion antigenicity. Virology 515, 191–202. 10.1016/j.virol.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 57.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep. 19, 719–732. 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z, Zhang Y, Cheng M, Ge N, Shu J, Xu Z, Su X, Kou Z, Tong Y, Qin C, and Jin X (2022). A single nonsynonymous mutation on ZIKV E protein-coding sequences leads to markedly increased neurovirulence in vivo. Virol. Sin. 37, 115–126. 10.1016/j.virs.2022.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu X, Yu F, Wu Y, and Ying T (2021). Potent germline-like monoclonal antibodies: rapid identification of promising candidates for antibody-based antiviral therapy. Antib. Ther. 4, 89–98. 10.1093/abt/tbab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu DC, Chumpolkulwong K, Corley MJ, Hunsawong T, Inthawong D, Schuetz A, Imerbsin R, Silsorn D, Nadee P, Sopanaporn J, et al. (2022). Neurocognitive impact of Zika virus infection in adult rhesus macaques. J. Neuroinflammation 19, 40. 10.1186/s12974-022-02402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim IJ, Lanthier PA, Clark MJ, De La Barrera RA, Tighe MP, Szaba FM, Travis KL, Low-Beer TC, Cookenham TS, Lanzer KG, et al. (2022). Efficacy of an inactivated Zika vaccine against virus infection during pregnancy in mice and marmosets. NPJ Vaccines 7, 9. 10.1038/s41541-021-00426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim IJ, Tighe MP, Clark MJ, Gromowski GD, Lanthier PA, Travis KL, Bernacki DT, Cookenham TS, Lanzer KG, Szaba FM, et al. (2023). Impact of prior dengue virus infection on Zika virus infection during pregnancy in marmosets. Sci. Transl. Med. 15, eabq6517. 10.1126/scitranslmed.abq6517. [DOI] [PubMed] [Google Scholar]
- 63.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng YQ, Dai JX, Ji GH, Jiang T, Wang HJ, Yang HO, Tan WL, Liu R, Yu M, Ge BX, et al. (2011). A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS One 6, e16059. 10.1371/journal.pone.0016059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271. 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saunders KO, Wang L, Joyce MG, Yang ZY, Balazs AB, Cheng C, Ko SY, Kong WP, Rudicell RS, Georgiev IS, et al. (2015). Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates from Mucosal Simian-Human Immunodeficiency Virus Infection. J. Virol. 89, 8334–8345. 10.1128/JVI.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goldenzweig A, Goldsmith M, Hill SE, Gertman O, Laurino P, Ashani Y, Dym O, Unger T, Albeck S, Prilusky J, et al. (2016). Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol. Cell. 63, 337–346. 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Calvet GA, Kara EO, Giozza SP, Bôtto-Menezes CHA, Gaillard P, de Oliveira Franca RF, de Lacerda MVG, da Costa Castilho M, Brasil P, de Sequeira PC, et al. (2018). Study on the persistence of Zika virus (ZIKV) in body fluids of patients with ZIKV infection in Brazil. BMC Infect. Dis. 18, 49. 10.1186/s12879-018-2965-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calvet GA, Kara EO, Landoulsi S, Habib N, Bôtto-Menezes CHA, Franca R.F.d.O., Neto AM, Castilho MDC, Fernandes TJ, Pereira GF, et al. (2021). Cohort profile: Study on Zika virus infection in Brazil (ZIKABRA study). PLoS One 16, e0244981. 10.1371/journal.pone.0244981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40. 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown EP, Licht AF, Dugast AS, Choi I, Bailey-Kellogg C, Alter G, and Ackerman ME (2012). High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods 386, 117–123. 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, et al. (2008). Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82, 12449–12463. 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davidson E, and Doranz BJ (2014). A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 143, 13–20. 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Commo FB, and B.M. (2016). Nplr: N-Parameter Logistic Regression. https://CRAN.R-project.org/package=nplr.
- 76.Dowd KA, DeMaso CR, Pelc RS, Speer SD, Smith ARY, Goo L, Platt DJ, Mascola JR, Graham BS, Mulligan MJ, et al. (2016). Broadly Neutralizing Activity of Zika Virus-Immune Sera Identifies a Single Viral Serotype. Cell Rep. 16, 1485–1491. 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 78.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- 79.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, and Eisenberg D (2006). Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103, 8060–8065. 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Henikoff S, and Henikoff JG (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are found within the paper and its Supplemental Information. The accession numbers for the Protein DataBank coordinates, and structure factors determined in this paper are PDB: 8EE8, 8EEE, 8EED, 8EE5, 8EEZ, 8EF1, 8EF0, 8EF2, and 8EF3, the GenBank accession numbers for the antibody sequences are GenBank: OR232903-OR232948.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


