Figure 6. Comparison of DENV and ZIKV antibody specificity.
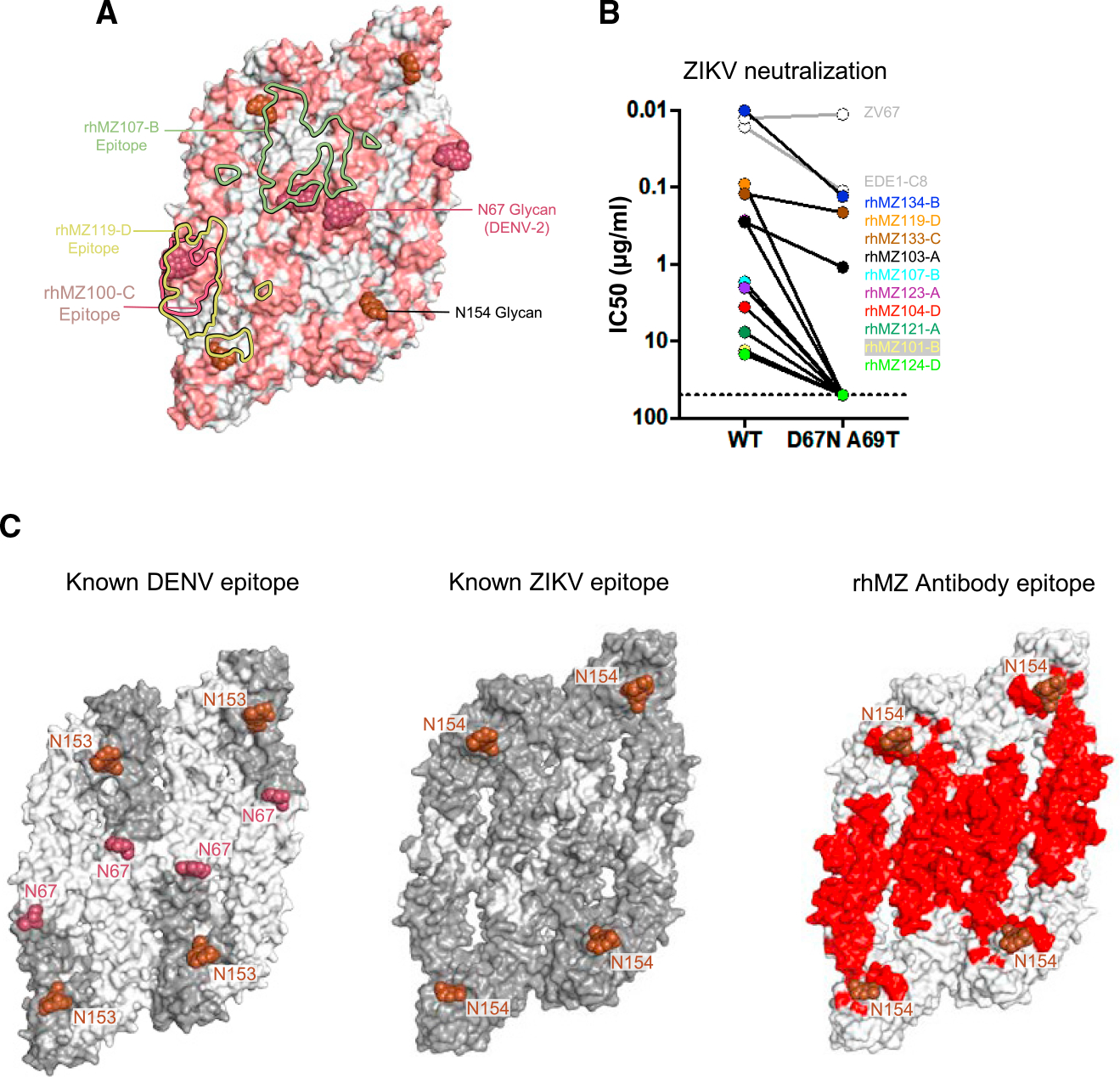
(A) Sequence differences between ZIKV E and DENV-2 E are mapped onto the ZIKV structure (PDB: 5IRE). Sequence and positional differences between ZIKV and DENV-2 are colored light red, while identical residues are colored white. Glycan-154 and DENV glycan-67 are shown in sphere representation colored brown and raspberry, respectively. The antibody binding footprints of rhMZ107-B, rhMZ100-C, and rhMZ119-D antibodies are shown as green, raspberry, and yellow solid lines, respectively.
(B) Neutralization (IC50, mg/mL) of wild-type (WT) and D67N-A69T mutant ZIKV performed in the ZIKV/H/PF2013 background using a reporter virus particle (RVP) assay. The addition of glycan-67 to ZIKV interfered with epitope recognition and abrogated or eliminated neutralization.
(C) Epitope mapping of structurally defined antibodies mapped onto four protomers of DENV (left) and ZIKV (middle). Residues contacted by previously described mAbs are colored dark gray, and residues not previously identified prior to this study are indicated in white. Only previously identified mAb structures with resolution greater than 4 Å were used since the contact residues are clearly interpretable. Newly identified residues contacted by rhMZ mAbs described in this study are colored red (right). Glycan sites at positions 153 or 154 are indicated in rose color.
See also Figures S5 and S7 and Table S6.
