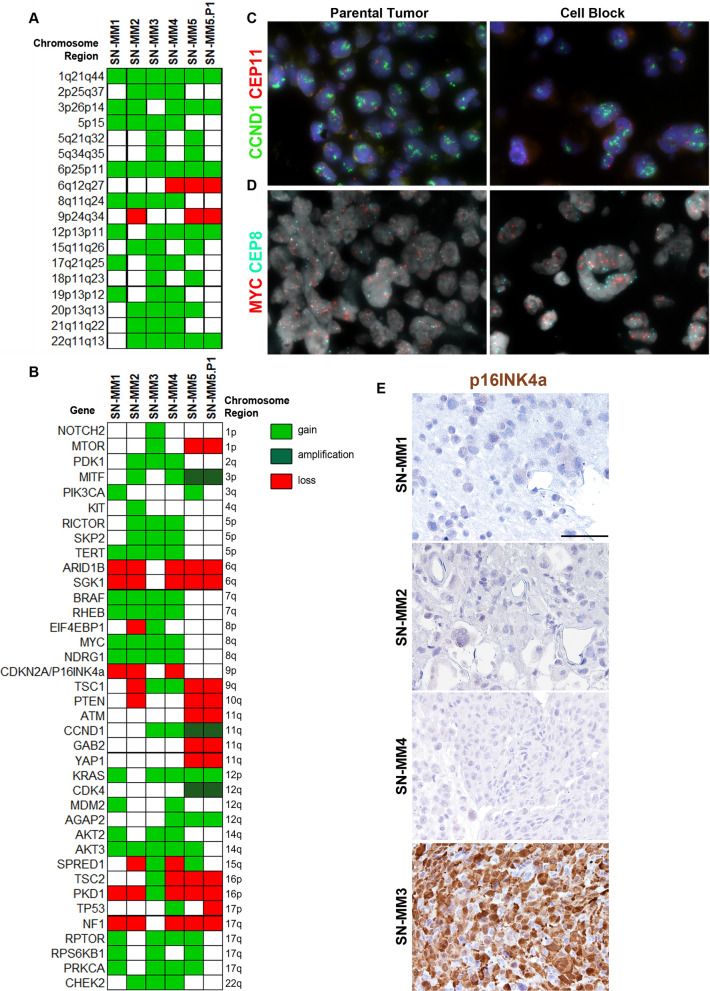
Fig. 3.
Significant chromosome regions and genes affected by copy number variation (CNV) in SN-MM cell lines. (A, B) Heatmaps show the chromosome regions recurrently affected by gains or losses (A) and significant genes, previously identified as melanoma driver genes or component of PI3K/Akt/mTOR pathway, affected by CNVs (B) as detected by aCGH. The green blocks indicate copy number gain/amplification and red blocks indicate copy number loss/deletion, as labeled. (C, D) FISH analysis of the amplified regions including CCND1 amplification in SN-MM5 (C) and MYC amplification in SN-MM3 (D) parental tumor biopsies and cell-blocks. (E) Immunohistochemistry for p16INK4a on cell blocks from SN-MM samples. Lack of p16INK4a expression is observed in SN-MM1, SN-MM2 and SN-MM4, whereas strong reactivity is observed in SN-MM3. Sections are counterstained with haematoxylin. Magnification 200X; scale bar 100 µm