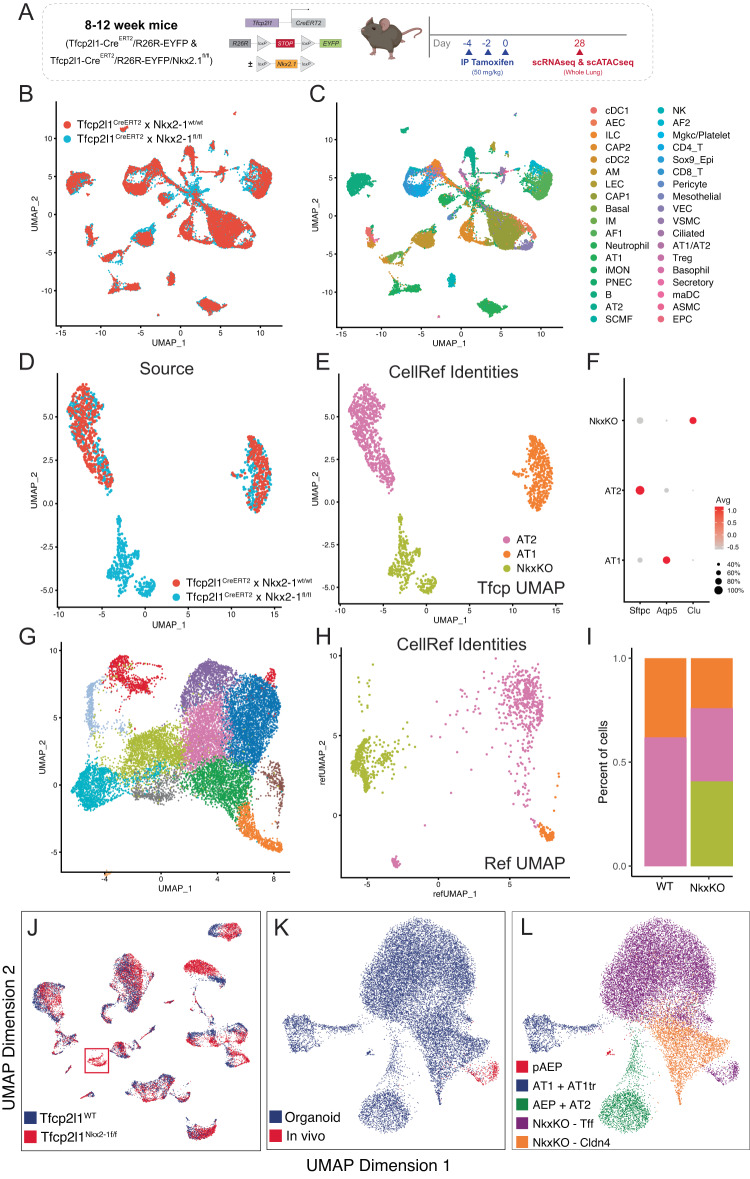
Fig. 8. In vitro and in vivo Nkx2-1 deletion generates a common PATS/Krt8+-like molecular state.
A Experimental design of in vivo genetic ablation of Nkx2-1 in AEPs prior to scRNAseq. B, C Common UMAP of whole-lung scRNAseq from WT and Nkx2-1KO animals. C shows cell identities using LungMAP labels. D–F Distal epithelial cell populations in WT (red) and Nkx2-1KO (blue) animals; a Nkx2-1KO specific population is present, which expresses markers of Krt8/PATS/DAPT+ cells (Clu) but not AT1 or AT2 cells. G–I Overlap and label transfer of AEP-O identities to in vivo epithelial cells from (D–F). G shows reference UMAP for comparison, reproduced from Fig. 6 with all NkxKO states concatenated together for comparison to in vivo cells. H shows clustering of in vivo epithelial cells on UMAP generated from organoids as in (G) and colored by identity as in (E). Proportions of cells in each state are shown in (I). J–L scATACseq comparison of Nkx2-1 knockout states in vivo and in vitro. J shows overall UMAP of single nuclear ATACseq from WT and Nkx2-1KO animals, demonstrating (as in RNA, compared to (D)) an Nkx2-1KO-specific epithelial population (red box). K Integrated scATAC analysis of overlap of Nkx2-1KO organoids (blue) and Nkx2-1KO–specific epithelial cluster from (J) (red). Cells comprising the Nkx2-1KO–specific epithelial cluster with Nkx2-1KO cells from AEP-O, confirming epigenomic similarity of the Nkx2-1KO cells generated in vivo and in vitro. Schematics created with Biorender.com.