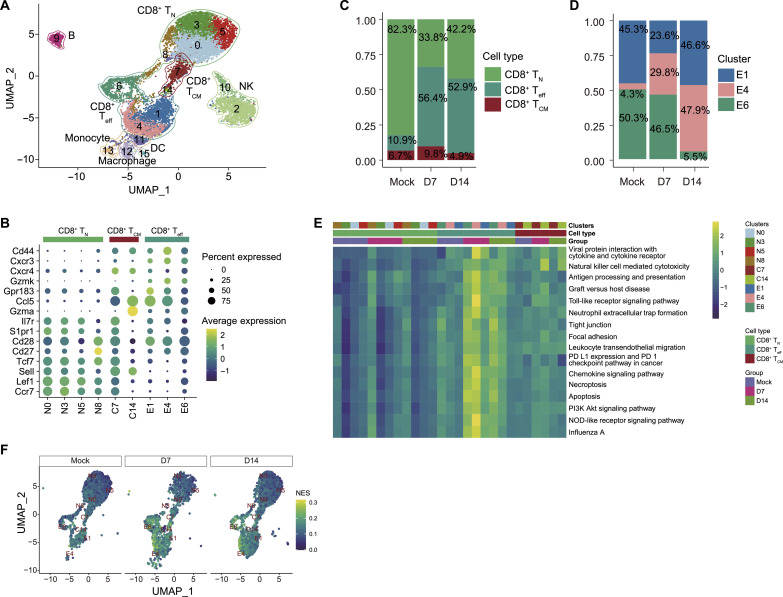
Fig. 2. Cellular and functional heterogeneity among CD8+ T cell subsets during peak influenza A virus (IAV) and post-IAV clearance phases.
Single-cell RNA-sequencing (scRNA-seq) analysis of FACS-sorted CD8+ T cells from mock- and PR8-infected mice lungs at 7 and 14 dpi. (A) Unbiased UMAP of total CD8+ T cells from mock- and PR8-infected mice lungs at 7 and 14 dpi, revealing 15 clusters of CD8+ T cells. (B) Dot plot representing expression levels of canonical marker genes for the selected 10 clusters generated by unbiased analysis and their corresponding subsets clustering within CD8+ TN, TCM, or Teff. Clusters were renamed as N for naïve, C for central, and E for effectors corresponding with their CD8+ T cell subsets. (C and D) Distribution of the proportions of CD8+ TN, TCM, and Teff clusters (C) and clusters E1, E4, and E6 (D), corresponding to the CD8+ Teff subset; and their kinetics at day 0 (mock), 7 (D7) and 14 (D14) dpi. (E) Heatmap of GSVA enrichment scores of selected significant pathways between CD8+ Teff against TN and TCM. Color stands for the level of up-regulation (yellow) or down-regulation (dark green) in cells. (F) GSVA enrichment scores of CD8+ T cells with UMAP embedding NK cell–mediated cytotoxicity pathway. Color stands for the normalized enrichment score (NES).