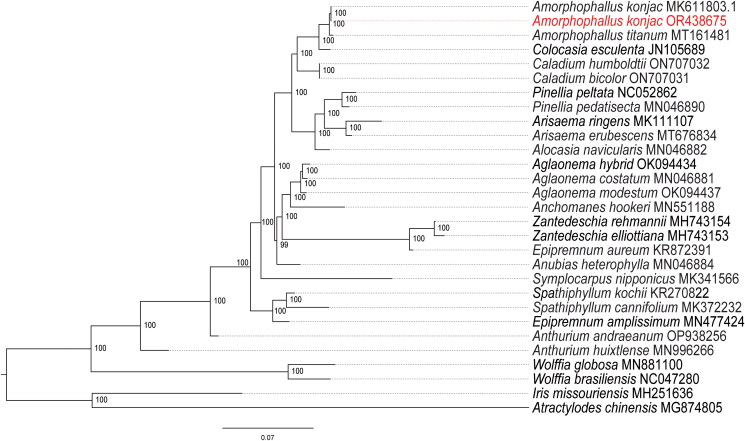
Figure 3.
Phylogenomic tree of Amorphophallus konjac (in red) and 28 species constructed using maximum-likelihood method based on complete chloroplast genome sequences. Iris missouriensis and Atractylodes chinensis are used as an outgroups. The best-fit model according to the bayesian information criterion (BIC) was TVM + F+R6. Numbers at each node represent the bootstrap values for 1000 replicates. The following sequences were used: Amorphophallus konjac MK611803.1 (Hu et al. 2019), Amorphophallus titanum MT161481 (Henriquez et al. 2021), Colocasia esculenta JN105689 (Ahmed et al. 2012), Caladium humboldtii ON707032 (Ye et al. 2022), Caladium bicolor ON707031 (Ye et al. 2022), Pinellia peltata NC052862 (unpublished), Pinellia pedatisecta MN046890 (Henriquez, Ahmed, et al. 2020), Arisaema ringens MK111107 (unpublished), Arisaema erubescens MT676834 (Zhang et al. 2020), alocasia navicularis MN046882 (Henriquez, Ahmed, et al. 2020), Aglaonema hybrid OK094434 (Li et al. 2022), Aglaonema costatum MN046881 (Henriquez, Ahmed, et al. 2020), Aglaonema modestum OK094437 (Li et al. 2022), anchomanes hookeri MN551188 (Henriquez, Ahmed, et al. 2020), Zantedeschia rehmannii MH743154 (He et al. 2020), Zantedeschia elliottiana MH743153 (He et al. 2020), Epipremnum aureum KR872391 (Tian et al. 2018), anubias heterophylla MN046884 (Henriquez, Ahmed, et al. 2020), symplocarpus nipponicus MK341566 (Kim et al. 2019), Spathiphyllum kochii KR270822 (Han et al. 2016), Spathiphyllum cannifolium MK372232 (Liu et al. 2019), Epipremnum amplissimum MN477424(unpublished), Anthurium andraeanum OP938256 (Wan et al. 2023), Anthurium huixtlense MN996266 (Henriquez, Mehmood, et al. 2020),Wolffia globosa MN881100 (unpublished), Wolffia brasiliensis NC047280 (unpublished), Iris missouriensis MH251636 (unpublished), Atractylodes chinensis MG874805 (Wang et al. 2020).