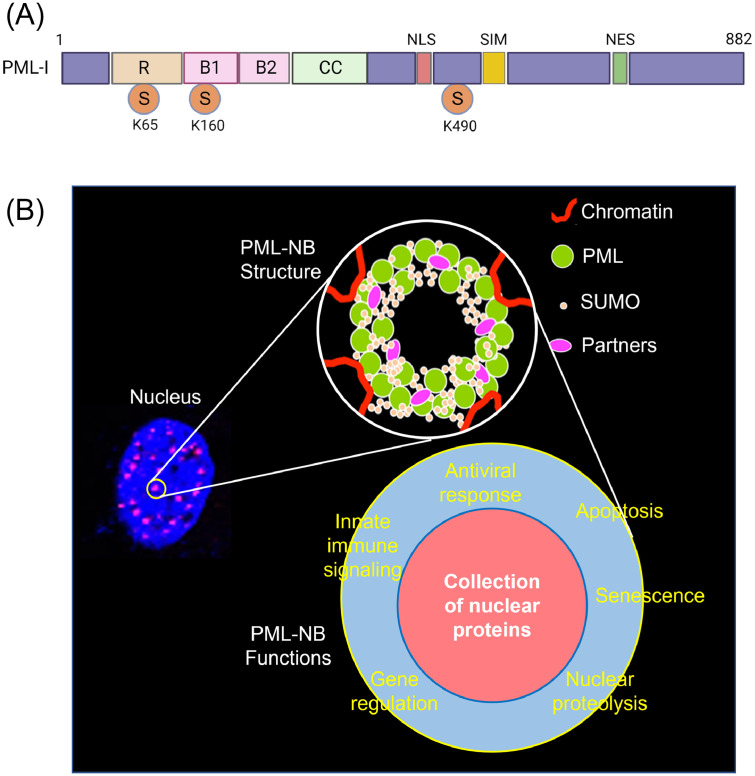
Fig. 1.
Structure and function of promyelocytic leukemia nuclear bodies (PML NBs). (A), Primary structure of the isotype I of PML. PML has a conserved structural arrangement of three domains (RBCC): RING (R) domain, two B-box (B1 and B2) domains, coiled-coil (CC) domain. The RING (really interesting new gene) domain possesses an E3-ubiquitin ligase activity which can conjugate ubiquitin to their targets as well as ubiquitin-like proteins such as ISG15 (interferon-stimulated gene 15) and SUMO (small ubiquitin-like modifier). Two B-box domains are thought to modulate protein-protein interactions. The CC domain mediates the homomeric or heteromeric assembly of PML proteins. PML is SUMOylated at three lysine residues of K65, K160, and K490. NLS, nuclear localization signal; SIM, SUMO-interacting motif; NES, nuclear export signal; S, SUMOylation sites (Adapted from Bernardi and Pandolfi, 2007). (B), PML NBs are dynamic membrane-less protein aggregates in the nucleus of the cell. PML NBs appear as dot-shaped spherical structures in the interchromatin nuclear space. PML is a key component for the structural integrity of PML NBs. PML NBs are formed initially by disulfide linkages of PML monomers as well as non-covalent interactions to drive the assembly of PML oligomerization. PML is SUMOylated and recruits other partner proteins to the inner core, and mature PML NBs are formed. PML NBs participate in a wide range of cellular processes such as cell cycle regulation, apoptosis, senescence, DNA repair, gene regulations, and antiviral response. Of these processes, antiviral response is the first line of intracellular defense against invading pathogens, and PML NBs can confer intrinsic immunity (Adapted from Hoischen et al., 2018).