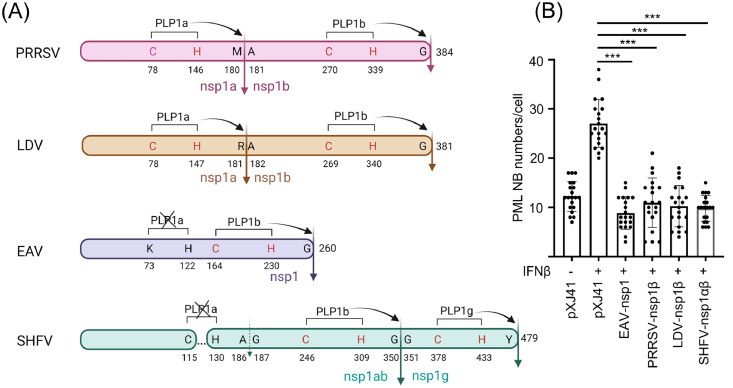
Fig. 5.
Reduction of PML NBs by nsp1 of different arteriviruses. (A), Biogenesis, structures, and cleavages of nsp1 proteins of arteriviruses. The full-length uncleaved nsp1 proteins for PRRSV, LDV, EAV, and SHFV are 384, 381, 260, and 479 amino acids (aa), respectively. The catalytic amino acid residues for PLPs are indicated in red. Solid vertical arrows represent the PLP-mediated cleavage sites. The dotted vertical arrow for SHFV indicates predicted cleavage site which is in fact uncleaved because PLP1α is inactive due to the deletion between 115 and 130 (dotted horizontal line). Crossing-outs indicate non-functional PLPs. Numbers indicate aa positions with respect to the full length of nsp1. The space between aa positions 115 and 130 in SHFV-nsp1 indicates a deletion in comparison with nsp1 of other arteriviruses (Adapted from Han et al., 2014). (B), Quantification of the reduction of PML-NBs in nsp1β-expressing cells. HeLa cells were transfected with indicated FLAG-tagged arterivirus nsp1 subunit genes for 24 h and stimulated with 1000 units/ml of IFN-β by incubation for 6 h. The cells were fixed and stained with anti-FLAG Ab (green) and anti-PML Ab (red) as described in Materials and Methods. Nuclei were stained with DAPI (blue). The images were taken by confocal microscopy (Nikon A1R). Twenty cells expressing nsp1 were randomly chosen, and numbers of PML-NB puncta per nucleus per cell were counted. The average numbers of PML NBs were calculated for each gene and used to construct the bar graphs. PLP, papain-like proteinase; PRRSV, porcine reproductive and respiratory syndrome virus; LDV, lactate dehydrogenase elevating virus of mice; EAV, equine arteritis virus; SHFV, simian hemorrhagic fever virus. ***, P<0.001. The figure was created with BioRender.com.