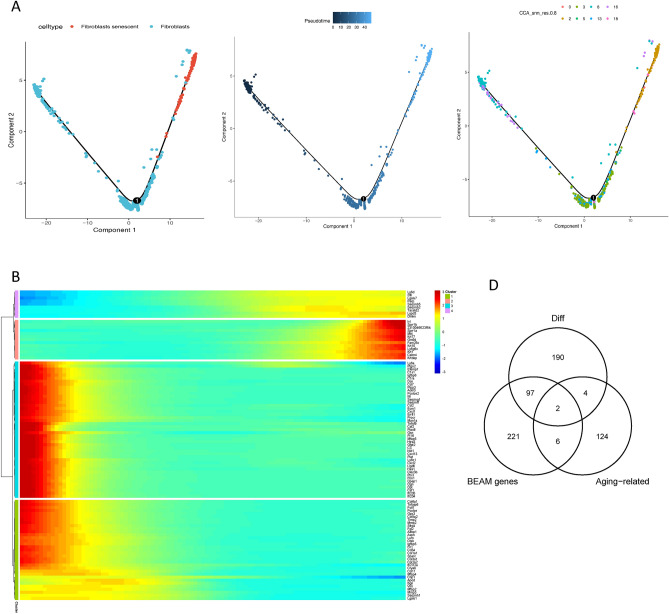
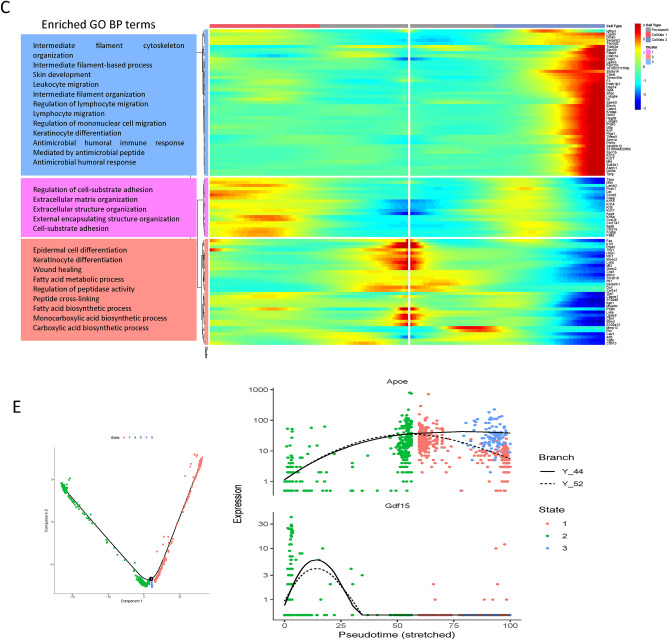
Figure 3.
Gene expression patterns differ as fibroblasts develop. (A) Trajectory diagram of Monocle after sorting of single cells. The horizontal coordinates indicate the two major components, each spot on the plot represents a cell, and the numbers in the black circles are the nodes in the trajectory analysis that define the various cell states. (The different colors in the left panel represent the different cell types, the middle panel is colored from dark to light in the proposed time order, and the different colors in the right panel represent the different cell Clusters) (B) Heat map of the top 100 genes that were expressed differently along pseudo-time (rows) and pseudo-time (columns), grouped hierarchically into four shapes. The genes are shown in the rows, and the probable time points are shown in the columns; the colors indicate the average expression levels of those genes at that moment; and the expression levels decrease from red to blue as time progresses. (C) The heat map shows the first 100 genes obtained from the analysis of the specified branch points in A. The top is the node minutes in the proposed time sequence, cell fate1 represents fibroblasts, and cell fate2 represents senescent fibroblasts. The left side of the heat map shows the GO function analysis for each cluster. (D) Genes linked with senescence and those differently expressed both at the pseudo-time nodes of the trajectory analysis and in senescent fibroblasts overlap in this Venn diagram. (E) Scatter plot of the expression pattern of Apoe, Gdf15 along the pseudo-time.