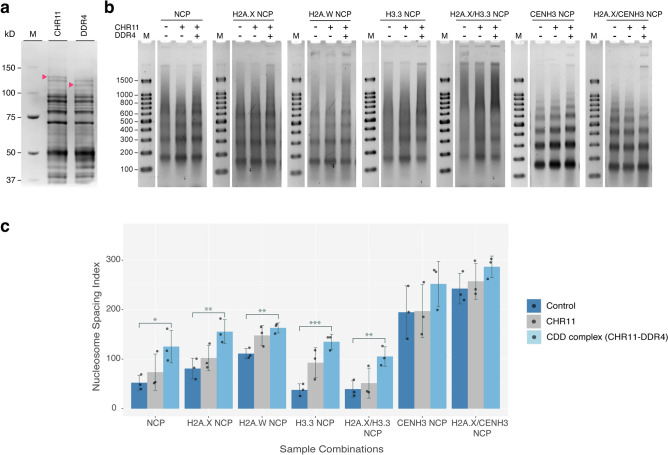
Figure 4.
The effect of ISWI remodeling complex on seven reconstituted chromatin combinations. (a) The cell-free synthesized CHR11 and DDR4 proteins are shown on a 10% SDS-PAGE, stained by CBB. (b) The partial MNase digestion assay of the seven chromatin combinations (NCP, H2A.X NCP, H2A.W NCP, H3.3 NCP, H2A.X/H3.3 NCP, CENH3 NCP, and H2A.X/CENH3 NCP) reacted in the absence of remodeling factors, the presence of CHR11 mRNA or the presence of both CHR11 and DDR4 mRNAs were analyzed on a 2% agarose gel. Uncropped agarose gel images and technical replicates for (b) are available in Supplementary Information (S4–S6). (c) Nucleosome spacing indexes (NSIs) of each combination were estimated from the agarose gel densitometry results of MNase assay (n = 3), and mean NSI values were plotted on a bar graph. The jitter plots over the bar graphs show the individual values of three replicas, the individual graphs of each replicate can be found in the Supplementary Information (S9). Error bars represent the standard deviation from the three independent experiments. Asterisks denote the statistical significance of NSI values between samples: *p value < 0.05; **p value < 0.01; ***p value < 0.001; non-significant results were not plotted.