Figure 3. Single cell BCR sequencing of SARS-CoV-2 antigen-specific B cells.
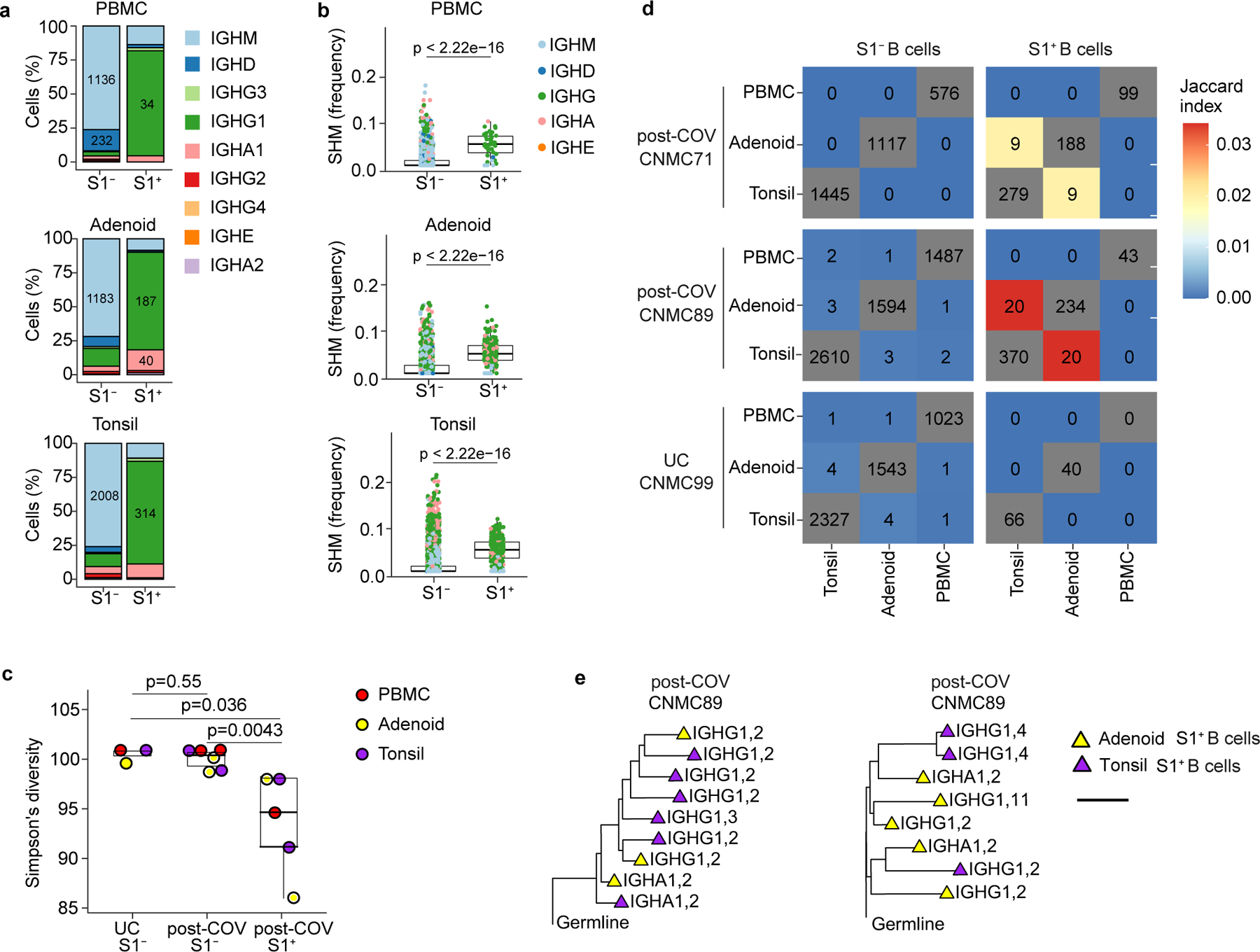
a. Sub-isotype percentages among sorted S1+ and S1− B cells from PBMC, adenoid and tonsil of one post-COV donor (CNMC89). Labels show the raw number of cells with a given sub-isotype and are only included for sub-isotypes that make up at least 10% of a given category. b. Somatic hypermutation (SHM) frequency among sorted S1+ and S1− B cells from PBMC, adenoid and tonsil of CNMC 89 (PBMC S1+ n= 44, S1− n=1491 cells; adenoid S1+ n=261, S1− n=1647 cells; tonsil S1+ n=416, S1− n=2644 cells). Mutation frequency calculated in V gene. Medians ± quartiles and p values are shown in the box plots. c. Simpson’s diversity of S1+ and S1− B cells from PBMCs, adenoids and tonsils from 2 post-COV donors (CNMC71 and 89) and S1− B cells from one UC (CNMC99). Lower Simpson’s diversity values indicate a greater frequency of large clones. d. Overlap of B cell clones among PBMCs, tonsils and adenoids from post-COV and UC donors. Off-diagonal elements are colored by the Jaccard index of clonal overlap between the two tissues and are labelled by the raw number of overlapping clones. Diagonal elements are labelled by the total number of clones within a particular tissue. e. Clonal lineage trees from two of the largest S1+ B cell clones shared between tonsil and adenoid from CNMC89. Triangles indicate S1+ cells, and tip color indicates tissue of origin (purple: tonsil; yellow: adenoid). Isotype and CITE-seq cluster of each cell are listed next to the symbol. Branch lengths represent SHM frequency/codon in VDJ sequence according to the scale bar. Significance calculated with two-sided Mann Whitney U test.
