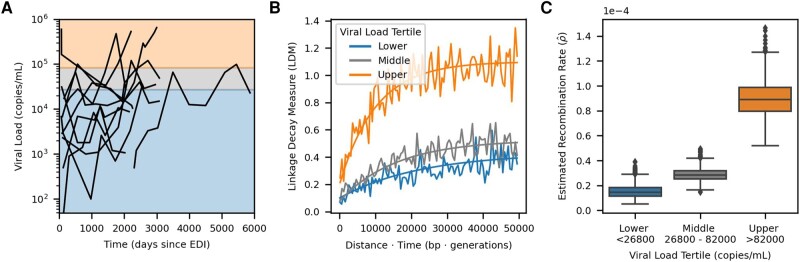
Fig. 4.
Elevated recombination rates are associated with high viral load in vivo. A) Viral load time courses for each participant are plotted. The background shading and horizontal demarcations indicate the viral load tertiles: (lower/blue), 26,800 to 82,000 (middle/gray), and (upper/orange) which divide the linkage decay measures (LDMs) into three approximately equal sized groups (≍ 20k LDMs per group). While viral loads at single time points are shown here, pairs of time points were used to form the groups for estimation and were sorted based on their average viral load (see supplementary Figs. S9 and S10, Supplementary Material online). B) Curve fits used by RATS-LD for estimation. Curve fits representing the median estimates of the bootstrap distribution (1,000 bootstraps) for each group are overlaid on the binned average of the LDMs from the in vivo data (moving average used for display purposes only, window size = 500 bp × generations). C) Recombination rate estimates with 1,000 bootstraps are shown separately for the three tertiles. Each box represents the quartiles of the distribution and the median is indicated by the central line.