Abstract
Gastrointestinal cancer is frequently detected at an advanced stage and has an undesirable prognosis due to the absence of efficient and precise biomarkers and therapeutic targets. Exosomes are small, living-cell-derived vesicles that serve a critical role in facilitating intercellular communication by transporting molecules from donor cells to receiver cells. circular RNAs (circRNAs) are mis-expressed in a variety of diseases, including gastrointestinal cancer, and are promising as diagnostic biomarkers and tumor therapeutic targets for gastrointestinal cancer. The main features of exosomes and circRNAs are discussed in the present review, along with research on the biological function of exosomal circRNAs in the development and progression of gastrointestinal cancer. It also assesses the advantages and disadvantages of implementing these findings in clinical applications.
Keywords: exosomes, circular RNA, gastrointestinal cancer, biomarker, therapeutic target
1. Introduction
Gastrointestinal cancer is among the most prevalent and predominant malignancies globally, consisting mainly of gastric cancer (GC) and colorectal cancer (CRC), accounting for 26% of the global cancer incidence and 35% of all cancer-associated mortalities in 2018 (1). At least ~1/3 (30–35%) of cases of gastrointestinal cancer are associated with imbalanced diet and lack of exercise; ~1/4 (20–30%) are associated with tobacco products; and ~1/5 (15–20%) are triggered by infectious agents (2).
Currently, fiberoptic gastroscopy and biopsy are the major invasive techniques used to diagnose gastrointestinal cancer (3). Not only can the diagnostic methods of gastrointestinal cancer be harmful, the diagnosis period of this disease is also mostly diagnosed at an advanced stage, which seriously affects the survival and prognosis of patients (3,4). The treatment of gastrointestinal cancer, which mostly involves the removal of the primary cancer tumor or the administration of medications such as cetuximab, paclitaxel and sorafenib, has likewise seen slow development in research (5). However, patients with surgically treated gastrointestinal cancer have some complications and a high recurrence rate, and patients treated with drugs will also develop a certain degree of drug resistance (5,6). Therefore, research into safer and more effective diagnostic and treatment approaches is urgently needed. Recent studies have found that targeted therapies are effective in treating certain types of cancers (7,8). In order to increase the survival rate of patients with gastrointestinal cancer, it is crucial to research innovative targeted molecules related to this illness.
Immune cells, stromal cells, extracellular matrix, substances released by cells (such growth factors and extracellular vesicles), lymphatic arteries, vascular networks and extracellular vesicles frequently exist in the tumor microenvironment (9). These elements interact in the tumor microenvironment to control the emergence, growth and metastasis of malignancies (9,10). Exosomes are small membranous vesicles that are produced when internal multivesicular bodies fuse with the cell membrane and are deposited in the extracellular matrix (11,12). A growing number of studies in recent years have revealed that exosomes can not only receive a wide range of neurotransmission and endocrine signaling, but also serve as a signaling platform for intercellular communication and coordinate various autocrine and paracrine functions, thereby influencing the microenvironment and development of tumors (13,14).
In addition to proteins, mRNAs, microRNAs (miRNAs/miRs) and circular RNAs (circRNAs) are also important components of exosomes (15). CircRNAs are abnormally expressed in cancer cells, as evidenced by the growing body of research, which has an impact on processes including cancer growth and treatment resistance (16,17). CircRNAs are stabler compared with other RNAs and vary from conventional linear RNAs in that they have a covalent closed-loop structure (18). Exosomal circRNAs have been shown to be useful as biomarkers for gastrointestinal cancer diagnosis, prognosis and treatment targets in a number of investigations (19,20). For example, Xing et al confirm that hsa_circ_0004831 is highly expressed in blood extracellular vesicles in patients with CRC. The results of this bioinformatics analysis identified miRNAs involved in the regulation of this circRNA, such as hsa-miR-4326, which may be related to the development of CRC (21).
The major traits and purposes of exosomes and circRNAs are reviewed in the present study, along with the biological significance and putative molecular pathways of exosomal circRNAs in the development of gastrointestinal cancer. Additionally, the present study provides an overview of the prospects and problems in this area and highlights novel developments in exosomal circRNAs as potential diagnostic biomarkers and therapeutic targets for gastrointestinal cancer.
2. Exosomes and circRNAs
Biogenesis, characteristics and functions of exosomes
Exosomes are a type of extracellular vesicle (EVs), while ectosomes are the other type (22). Compared with ectosomes, the biological origin and particle diameter of exosomes are different. After forming an outward bud, ectosomes sever the surface of the plasma membrane, and their diameters range from 50 nm to 1 mm (23). However, exosomes are nanoscale vesicles originating from endosomes with diameters of 40–160 nm (average, ~100 nm) (23). All cells secrete exosomes in normal and pathological conditions, which maintain cell homeostasis and mediate intercellular communication (24). Exosomes contain numerous cellular components, including nucleic acids, amino acids, lipids, glycans, metabolites, cytoplasm and cell surface proteins (24). In exosomes from distinct cell types, 9,769 proteins, 3,408 mRNAs, 2,838 miRNAs and 1,116 lipids have so far been discovered (25). Exosomes have been the subject of an increasing number of research in recent years due to their potential use as non-invasive liquid biopsy instruments for the detection and treatment of a variety of disorders as well as their potential for usage in prescription drugs (23,26).
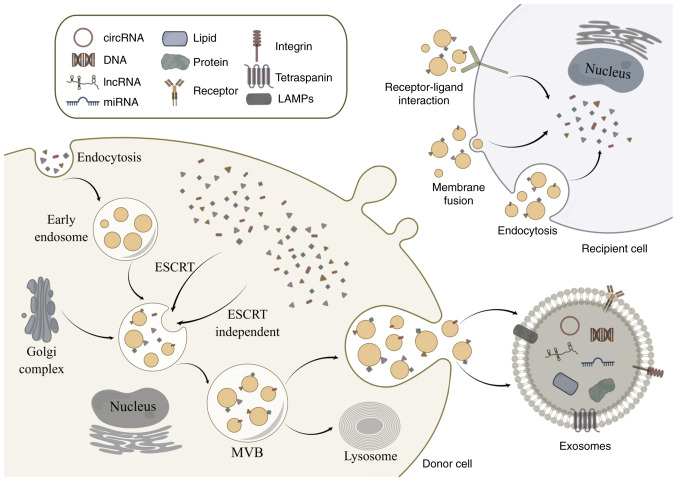
Exosomes were discovered for the first time in sheep reticulocytes by Pan and Johnstone in 1983 (27). Initially considered as cellular debris, exosomes were later found to have independent structures (27). The occurrence of exosomes starts from the vesicles on the cell membrane, which turn inward to form exosomes, and the cytoplasmic membrane invaginates to enclose some specific extracellular biological macromolecules and cell membrane proteins to form intracellular multivesicular bodies (MVBs) (28). After the formation of MVBs, cells may fuse with autophagosomes or lysosomes, during which the previously encapsulated substances may be degraded or released through the fusion of the endosome with the plasma membrane to form exosomes (29). At present, several possibilities have been suggested for cargo sorting into exosomes, but the specific mechanism remains unclear (30). Exosome release and biomolecule packing into exosomes are reportedly regulated by the endosomal sorting complex required for transport (ESCRT) pathway, and ESCRT proteins import ubiquitin proteins into MVBs (30). A total of five protein-protein complexes make up the ESCRT system: i) ESCRT-0; ii) ESCRT-I; iii) ESCRT-II; iv) ESCRT-III; and v) Vps4-Vta1 (31). In summary, ESCRT-0 gathers ubiquitinated cargo to start the process; protein complexes in a saddle shape are created when the ESCRT-I and ESCRT-II complexes bind cargo; the ESCRT-III complex promotes vesicle contraction and maturation; and Vps4-Vta1 promotes membrane fracture (32). In addition, some ESCRT-independent pathways also play important roles in exosome genesis. Lipid rafts, such as flotillins and caveolins, play a key role in ESCRT-independent intraluminal vesicles formation (33,34). Exosome production has also been linked to flotillin, caveolin-1, cholesterol and tetraspanins (Fig. 1) (35–38).
Figure 1.
Biogenesis of exosomes. Exosomes are derived from the fusion of MVBs. Exosomes can transport nucleic acids, proteins and lipids to recipient cells from doner cells to recipient cells mainly through receptor-ligand interaction, membrane fusion and endocytosis. By Figdraw (https://www.figdraw.com/#/). MVB, multivesicular body; circRNA, circular RNA; lncRNA, long non-coding RNA; miRNA, microRNA; ESCRT, endosomal sorting complex required for transport.
As aforementioned, exosomes were initially considered waste products excreted by cells. However, exosomes can control the proliferation, migration and invasion of tumor cells by information transfer, as more and more studies have demonstrated (39,40). For instance, He et al revealed that pancreatic cancer cells release exosomes that are abundant in FGD5 antisense RNA 1, which facilitates the evolution of pancreatic cancer by way of tumor-associated alternatively activated (M2) macrophage polarization (41). Exosomes have also been revealed to modulate immune responses (42). According to Yang et al, exosomal hsa_circ_0085361 is abnormally expressed in bladder cancer tissues and controls T cell depletion by influencing downstream miRNAs (43). Exosomes can also be crucial in a number of biological processes of the neurological system (44). According to studies, damaging proteins such β amyloid peptide, superoxide dismutase and α synuclein are present in the exosomes of the central nervous system and help advance disorders (44–47). For example, studies have found that α synuclein, which is secreted by cells through exosomal calcium-dependent mechanisms, helps amplify and spread Parkinson's disease-related pathology (47). Exosomes also contribute to the development of metabolic and other disorders, such as cardiovascular ailments (23). Notably, there are several potential uses for exosomes in the treatment, identification and assessment of tumors. According to Xu et al, the exosome-related gene fibroblast growth factor 9 can be downregulated and exploited as a novel diagnostic and prognostic target for ovarian cancer (48). Because a number of components of exosomes are related to diseases, research on exosomes has increased (49,50).
Properties and functions of circRNAs
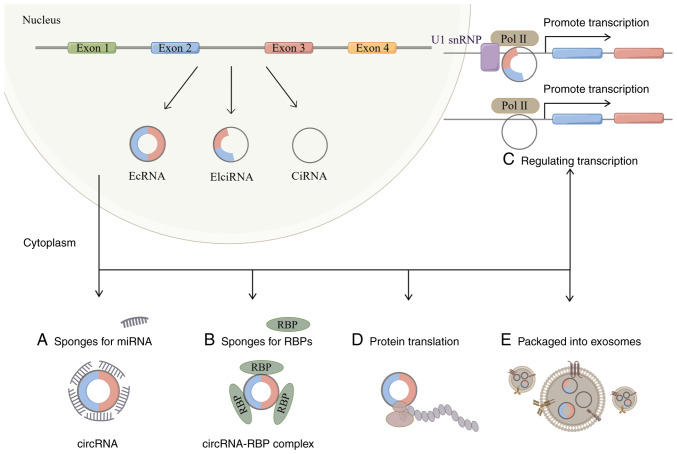
CircRNAs are closed circular RNA molecules found in a variety of specimen types, including bone marrow cells, platelets, white blood cells and red blood cells (51). Unlike conventional linear RNA, circRNA backsplicing joins the upstream 3′ splice acceptor site and the downstream 5′ splice donor site to create a single-stranded covalent closed loop (52). Circular RNA is more durable compared with linear RNA and resistant to exonuclease disintegration because it lacks the 5′ and 3′ ends (52). According to the various genome constituent sequences, circRNAs can be divided into three different categories: Exonic circular RNAs, intronic circular RNAs (ciRNAs) and exon-intron circular RNAs (EIciRNAs) (Fig. 2) (53). The majority of them are present in the nucleus, including ciRNAs and EIciRNAs; however, there are few miRNA targets for ciRNAs (53,54).
Figure 2.
Biogenesis and function of circRNAs. CircRNAs are generated by a back-splicing process that ligates a downstream 5′ splice donor site with an upstream 3′ splice acceptor site to form a single-strand covalently closed loop. Then, the spliceosome removes all or part of introns and the rest of sequences are connected, generating three types of circRNAs: EcRNAs, EIciRNAs and CiRNAs. CircRNAs have numerous functions, such as binding to miRNAs and RBPs, regulating transcription, promoting protein translation and being packaged into exosomes to come into play. By Figdraw (https://www.figdraw.com/#/). circRNA, circular RNA; EcRNAs, exonic circular RNAs; ciRNAs, intronic circular RNAs; EIciRNAs, exon-intron circular RNAs; Pol, polymerase; snRNP, small nuclear ribonucleoproteins; miRNA, microRNA; RBPs, RNA binding proteins. A, CircRNAs bind to miRNAs; B, CircRNAs bind to RBPs; C, CircRNAs promote transcription; D, CircRNAs promote protein translation; E, CircRNAs are packaged into exosomes.
Due to its distinctive structure, circRNAs have a wide range of biological roles and are crucial in numerous illnesses, according to recent studies (55,56). CircRNAs can control gene expression by interacting with miRNAs, as a number of studies have demonstrated (57,58). For example, Huang et al has revealed that hsa_circ_104348 can regulate the expression of miR-187-3p, thereby affecting the progression of hepatocellular carcinoma (59). The exonic region of RNase P RNA component H27 is the source of circRNA-002178, which has been shown to be overexpressed in esophageal cancer cells (60). CircRNA-002178 can act as a sponge for miR-34a and miR-28-5p to reduce their inhibitory effects on their target genes PDL1 and PD1 in cancer cells and CD8 T cells (60,61). Circular RNAs can also function by attaching to RNA-binding proteins. A new circRNA has been identified by Chen et al in the friend leukemia virus integration 1 promoter, and it is reported to encourage breast cancer spread by controlling the enzymes methylcytosine dioxygenase TET1 and DNA methyl transferase (62). Due to the absence of the 5′ cap, it has also been proposed that circRNAs translate in a cap-independent manner (54,63).
At present, the following circRNAs that can be translated into proteins have been studied: circ-AKT3 (64), circSHPRH (65) and circ-ZNF609 (66). For example, circ-AKT3 is expressed at a lower level in glioblastoma tissue compared with normal brain tissue, and it can encode AKT3-174aa (64). The tumorigenicity of glioblastoma can be affected by influencing the expression of circ-AKT3 and AKT3-174aa (64). As circRNA research advances, these types of molecules may be employed as tumor diagnostic markers. For example, Lu et al demonstrated that hsa_circ_0001789 is underexpressed in GC tissues and plasma, with an area under receiver operating curve (AUC) of 0.82; this circRNA may become a novel diagnostic biomarker for GC, and its efficacy is better compared with that of traditional tumor markers (67). According to a previous study, the hepatocellular carcinoma diagnostic biomarkers hsa_circ_0004001, hsa_circ_0004123, hsa_circ_0075792 and the combination of the three exhibit greater sensitivity and specificity compared with the three circRNAs separately (68).
3. Biological functions of exosomal circRNAs in gastrointestinal cancer
Role of exosomal circRNAs in gastrointestinal cancer
According to previous studies, circRNAs influence various biological processes, are persistent and are concentrated in exosomes (69,70). Exosomal circPRRX1, as the competing endogenous RNA of miR-596, has been revealed to promote the proliferation, migration and invasion of GC cells, and decreases their radiation sensitivity through the upregulation of NF-κB-activated protein (71). Since a number of cell types can receive exosomes, exosomes can act as messengers in intercellular communication (72). In the occurrence and development of tumors, circRNA can be packaged into exosomes and sent to receptor cells to play a certain role, affecting the tumor microenvironment and thus influencing the progression of the disease (73). As shown in Fig. 1, exosomes transfer circRNAs into recipient cells mainly via three processes: Receptor-ligand interaction, membrane fusion and endocytosis (23). However, the specific mechanism of selective packaging of circRNAs into exosomes is still unclear. According to Yang et al, the exosome circ-133 produced by hypoxia cells is substantially expressed in the plasma of CRC patients and can be transferred to healthy cells to advance the disease (74). Notably, numerous other studies have found a connection between exosomes and the removal of intracellular circRNAs (75,76). For example, it has been revealed that exosomes can be further eliminated by the reticuloendothelial system or produced by the kidney and liver (75). Moreover, previous research has revealed that, compared with healthy cells, CRC cells can actively release more circ-rho-related BTB domain-containing 3 (circRHOBTB3) to sustain colorectal cancer cell fitness (76). In another study, Chen et al demonstrated that circRHOBTB3 inhibits the progression of CRC by regulating intracellular reactive oxygen species and metabolic pathways (76). In addition, this study advanced a novel theory of tumor escape known as ‘the tumor exosome escape mechanism’, which postulates that tumor cells secrete tumor inhibitory circRNAs in order to maintain the fitness of cancer cells. The mechanism of tumor exosome escape is of great significance for understanding and treating cancer (76). Furthermore, the expression levels of circRNA in some patients with tumors are significantly different compared with that of the healthy population, such as hsa_circ_001783 and circRNA_102231, and because of its strong stability, long half-life and anti-degradation, circRNA can be used as a tumor diagnosis and molecular marker (77,78).
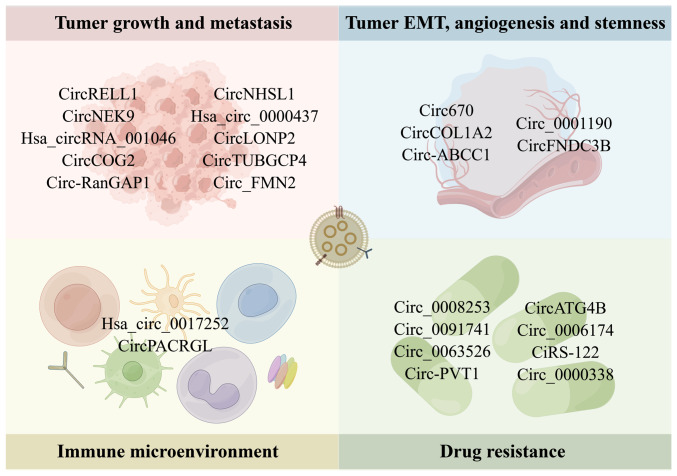
Exosomal circRNAs have been implicated in the emergence and progression of gastrointestinal cancer in a growing number of studies over the past several years (79,80). By binding to miRNA, receptor proteins, or other regulatory mechanisms, exosomal circRNAs influence tumor growth, metastasis, stemness, angiogenesis, the immunological microenvironment and treatment resistance (Fig. 3 and Table I).
Figure 3.
Biological role of exosomal circRNAs in gastrointestinal cancer. Exosomal circRNAs are involved in the tumor growth, metastasis, EMT, angiogenesis, stemness, immune microenvironment and drug resistance of gastrointestinal cancer. By Figdraw (https://www.figdraw.com/#/). circRNA, circular RNA; EMT, epithelial-mesenchymal transition.
Table I.
Function and mechanism of exosomal circRNAs in gastrointestinal cancer.
| Cancer type | CircRNA | CircBase ID | Function | Mechanism | (Refs.) |
|---|---|---|---|---|---|
| GC | CircRELL1 | Hsa_circ_0001400 | Modulating progression | MiRNA sponge for miR-637 | (82) |
| GC | CircNEK9 | Hsa_circ_0032683 | Accelerating the progression | Regulating miR-409-3p/MAP7 axis | (83) |
| CRC | Hsa_circRNA_001046 | Hsa_circ_0000395 | Promoting the progression | Elevating MYH9 expression by sequestering miR-432-5p | (84) |
| CRC | CircCOG2 | Hsa_circ_0016866 | Promoting the progression | Regulating miR-1305/TGF-β2/SMAD3 pathway | (85) |
| GC | Circ-RanGAP1 | Hsa_circ_0063526 | Facilitate invasion and metastasis | MiRNA sponge for miR-877-3p to regulate VEGFA expression | (86) |
| GC | CircNHSL1 | Hsa_circ_0006835 | Impeded migration, invasion, and glutaminolysis | Regulating miR-149-5p/YWHAZ axis | (87) |
| GC | Hsa_circ_0000437 | Hsa_circ_0000437 | Regulating proliferation, invasion, migration and apoptosis | Targeting SRSF3 and inhibiting PDCD4 | (88) |
| CRC | CircLONP2 | Hsa_circ_0008558 | Enhancing invasion and metastasis | Modulating the maturation and exosomal dissemination of miRNA-17 | (89) |
| CRC | CircTUBGCP4 | Hsa_circ_0101501 | Promoting vascular endothelial cell tipping and tumor metastasis | Activating Akt signaling pathway | (90) |
| CRC | Circ_FMN2 | Hsa_circ_0005100 | Regulating the proliferation, metastasis and apoptosis | Regulating miR-338-3p/MSI1 axis | (91) |
| GC | Circ670 | Hsa_circ_0000670 | Promoting the spheroidizing ability, stemness genes expression and EMT | Inducing the expression of circ670 | (95) |
| CRC | CircCOL1A2 | Hsa_circ_0081069 | Inhibiting proliferation, migration, invasion and EMT | Regulating miR-665/LASP1 signal axis | (96) |
| CRC | Circ-ABCC1 | Hsa_circ_0000677 | Regulating stemness and metastasis | Carrying circ-ABCC1 by exosomes from CD133 cells | (97) |
| GC | Circ_0001190 | Hsa_circ_0001190 | Inhibiting proliferation and angiogenesis | Regulating miR-586/SOSTDC1 axis | (98) |
| CRC | CircFNDC3B | Hsa_circ_0006156 | Regulating growth, metastasis and angiogenesis | Sequestrating miR-937-5p to derepress TIMP3 | (99) |
| GC | Hsa_circ_0017252 | Hsa_circ_0017252 | Attenuating progression | Suppressing macrophage M2-like polarization | (100) |
| CRC | CircPACRGL | Has_circ_0069313 | Promoting differentiation of N1 to N2 neutrophils | Regulating miR-142-3p/miR-506-3p-TGF-β1 axis | (101) |
| GC | Circ_0008253 | Hsa_circ_0008253 | Enhancing OXA resistance | Transferring from tumor-associated macrophage to gastric carcinoma cells | (103) |
| GC | Circ_0091741 | Hsa_circ_0091741 | Promoting cell autophagy and chemoresistance | Regulating miR-330-3p/TRIM14/Dvl2/Wnt/β-catenin axis | (104) |
| GC | Circ_0063526 | Hsa_circ_0063526 | Enhancing cisplatin resistance | Regulating the miR-449a/SHMT2 axis | (105) |
| GC | Circ-PVT1 | Has_circ_0009143 | Enhancing cisplatin resistance | Regulating miR-30a-5p/YAP1 axis | (106) |
| CRC | CircATG4B | Hsa_circ_0007159 | Inducing oxaliplatin Resistance | Promoting autophagy | (107) |
| CRC | Circ_0006174 | Hsa_circ_0006174 | Contributing to the chemoresistance of doxorubicin | Depending on the miR-1205/CCND2 axis | (108) |
| CRC | CiRS-122 | Hsa_circ_0005963 | Inducing chemoresistance | Regulating miR-122-PKM2 axis | (109) |
| CRC | Circ_0000338 | Hsa_circ_0000338 | Enhancing 5-fluorouracil resistance | Regulating miR-217 and miR-485-3p | (110) |
GC, gastric cancer; CRC, colorectal cancer; circRNA, circular RNA; MAP7, microtubule-associated protein 7; MYH9, myosin heavy chain 9; TGF-β, transforming growth factor beta; SMAD3, recombinant SMAD family member 3; VEGFA, vascular endothelial growth factor A; YWHAZ, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta; SRSF3, ser/arg-rich splicing factor 3; PDCD4, programmed cell death 4; Akt, protein kinase B; MSI1, musashi-1; EMT, epithelial-mesenchymal transition; LASP1, LIM and SH3 protein 1; SOSTDC1, sclerostin domain containing 1; TIMP3, tissue inhibitor of metalloproteinase 3; OXA, oxaliplatin; TRIM14, tripartite motif containing 14; DVL2, disheveled segment polarity protein 2; SHMT2, serine hydroxymethyltransferase 2; YAP1, yes-associated protein 1; CCND2, cyclin D2; PKM2, The M2 isoform of pyruvate kinase.
Roles of exosomal circRNAs in gastrointestinal tumor proliferation, metastasis, stemness and angiogenesis
Exosomal circRNAs have been shown to control the microenvironment and surrounding tissue cells, which has been demonstrated to have an impact on how gastrointestinal cancer progresses (81). It has been reported that exosomal circRNAs promote the occurrence and development of gastrointestinal cancer by affecting tumor proliferation, metastasis, stemness and angiogenesis (81). Sang et al demonstrated that circRNA RELL1 is significantly downregulated in GC tissues and transmitted by exosomes (82). CircRELL1 inhibits GC progression through the miR-637/ephrin type-B receptor 3 axis, which may be a novel diagnostic marker and therapeutic target for GC (82). Yu et al revealed that circRNA NIMA-related kinase 9 accelerates GC progression by targeting the miR-409-3p/microtubule associated protein 7 axis, which can be reflected in malignant behaviors such as migration and invasion (83). A possible therapeutic target for CRC has been identified in research as the cancer-derived exosomal hsa_circRNA_001046, which plays an oncogenic role in the progression of the disease (84). According to Gao et al, circRNA component of oligomeric Golgi complex 2 (COG2) may be transported from cancer cells with a high metastatic potential to cancer cells with a low metastatic potential via exosomes (85). circCOG2 has been demonstrated to be enhanced in CRC tissue and plasma exosomes. By controlling downstream miRNA and associated proteins, circCOG2 aids in the development of the malignant phenotype of CRC. According to this, circCOG2 may be both a therapeutic target and a predictive factor for CRC (85).
In addition, there is growing evidence that exosomes released by various living cells promote the metastasis of gastrointestinal cancer by delivering circRNAs (86,87). For example, circ-RanGAP1 is found in plasma exosomes from preoperative patients with GC, according to the research of Lu et al (86). In addition, the ability of GC cells to migrate and invade is improved by the plasma exosomes that were obtained from these individuals. This research raises the possibility that circ-RanGAP1 might serve as a predictive biomarker and a therapeutic target for the treatment of GC (86). Hui et al demonstrated that circNHSL1 knockdown represses migration, invasion and glutaminolysis and inhibits tumor growth by downregulating the miR-149-5p/tyrosine 3-monooxygenase axis in GC, implying an underlying therapy target for GC treatment (87). Shen et al also revealed that hsa_circ_0000437 regulates malignant behaviors such as migration of human lymphatic endothelial cells by targeting serine and arginine rich splicing factor 3 and inhibiting programmed cell death factor 4, as well as lymph node metastasis (LNM) in the popliteal LNM model (88). In CRC, circLONP2 is abnormally highly expressed in metastatic primary CRC tissues and can be transferred between cells by regulating miR-17 (89). This study suggests that circLONP2 may be a potential anti-metastatic therapeutic target for CRC (89). The findings of Chen et al determine that CRC cells are rich in exosomal circTUBGCP4, which promotes CRC metastasis through upregulation of pyruvate dehydrogenase kinase 2 (90). Additionally, Yu et al revealed that the exosomal circ_FMN2 is abundant in CRC cells and that its overexpression can encourage the spread of CRC cells and the development of colorectal tumors (91).
The incidence, invasion, metastasis and treatment resistance of gastrointestinal cancer are all assumed to be influenced by epithelial-mesenchymal transition (EMT), a fundamental cellular process (92). CircRNAs are crucial to EMT in gastrointestinal cancer (93,94). Exosomal circ670 from the tissues of patients with GC has been demonstrated to alter the EMT process of GC stem cells, hence boosting the growth of GC, according to a previous study (95). According to Miao et al, exosomal circRNA collagen type I α 2 chain (circCOL1A2) is significantly expressed in CRC cells, and miR-665 can impact EMT and other cellular properties when combined with circCOL1A2 or LIM and SH3 Protein 1 (96). Moreover, this study indicated that circCOL1A2 is a potential novel therapeutic target for CRC (96).
Additionally, it has been shown that a number of exosomal circRNAs are associated with the stemness and angiogenesis of gastrointestinal tumor cells (97). Zhao et al demonstrated that the exosomes of CD133 cells are rich in hsa_circ_0000677, which can mediate the stemness of CRC cells and promote the progression of CRC, and is expected to become a potential therapeutic target for this cancer (97). Liu et al also revealed that the expression of circ_0001190 in the exosome source of GC is low, and overexpression of circ_0001190 can inhibit the angiogenesis ability of cells, thus inhibiting the development of GC (98). Additionally, Zeng et al demonstrated that circFNDC3B has a low expression level in CRC tissues and cells and that it has the ability to stop the growth of the disease by preventing the angiogenic features of the disease (99). These studies indicate that exosomal circRNAs can promote or inhibit the progression of gastrointestinal cancer.
Role of exosomal circRNAs in the gastrointestinal tumor immune microenvironment
Exosomal circRNAs can affect the function of immune cells in the tumor microenvironment and then affect the occurrence and development of gastrointestinal cancer. For instance, exosomal hsa_circ_0017252 secreted by GC cells slows the growth of the disease by inhibiting M2-like polarization of macrophages (100). Exosome-derived circPACRGL is significantly expressed in CRC cells. By controlling downstream miRNAs, circPACRGL promotes the malignant activity of CRC cells and the differentiation of N1 to N2 neutrophils. To the best of our knowledge, this research is the first to demonstrate that exosomal circPACRGL contributes to CRC carcinogenesis and may have promise as a biomarker (101). These findings suggest that exosomal circRNAs can affect the progression of gastrointestinal cancer through immune cells.
Enhanced roles of exosomal circRNAs in drug resistance of gastrointestinal cancer
It has been revealed that circRNAs cannot only affect the progression of gastrointestinal cancer, but also participate in their chemoresistance (102). A number of circRNAs have a role in drug resistance in GC, such as circ 0008253 and circ_0091741 (103,104). According to a recent study, circ 0008253 is abundant in M2-polarized exosomes that are produced by macrophages and can be delivered to GC cells via exosomes to increase their resistance to oxaliplatin (OXA) (103). Chen et al also demonstrated that the exosomal circ_0091741 derived from GC cells increases the expression of tripartite motif containing 14, thereby enhancing OXA resistance (104). In addition, circ_0063526 is highly expressed in GC tissues and cells, and cisplatin resistance is enhanced by regulating the expression of serine hydroxymethyltransferase 2 (105). Moreover, exosome-derived circ-PVT1 is underexpressed in GC cells and serum, and contributes to DDP resistance by regulating miR-30a-5p; therefore, Exosomal circ-PVT1 may be a potential therapeutic target for GC (106). In CRC, a novel support for a possible therapeutic target for oxaliplatin resistance is provided by the exosomal circATG4B, which contributes to the lower chemosensitivity of CRC cells (107). Zhang et al revealed that circ_0006174 is upregulated in exosomes of doxorubicin (DOX)-resistant CRC cells and can enhance their resistance through the intercellular transfer of exosomes (108). Another study has revealed that hsa_circ_0005963 can be transferred from chemotherapy-resistant CRC cells to sensitive cells via exosomes, enhancing drug resistance in sensitive cells (109). A study suggests that hsa_circ_0005963 may be a potential therapeutic target for drug-resistant CRC (109). Moreover, Zhao et al revealed that miR-217 and miR-485-3p are controlled by exosome-mediated circ_0000338 transfer, which increases 3-fluorouracil resistance in CRC (110). These results suggest that exosomal circRNAs can enhance the resistance of oxaliplatin, 3-fluorouracil, cisplatin and DOX in gastrointestinal cancer. The application of antitumor drugs and specific circRNA inhibitors can improve the therapeutic effect of gastrointestinal tumors.
4. Exosomal circRNAs as potential biomarkers of gastrointestinal cancer
Because most of the early symptoms of gastrointestinal cancer are not obvious, early detection and diagnosis are challenging, and most patients are at an advanced stage when they have obvious symptoms, and their survival rate is low (111). Therefore, the search for new biomarkers for gastrointestinal cancer is crucial. Exosomes exist stably in body fluids and can act as messengers between cells, which are considered as promising biomarkers (Fig. 4 and Table II) (112,113). CircRNAs have been found to be stable and abundant in exosomes, making it simple to detect their levels (114,115). In addition, circRNAs can be specifically and differentially expressed across tissues and body fluids under various pathological conditions, so circRNAs can be used as a novel biomarker for a number of diseases (116,117). A previous study has shown that the circulating levels of exosomal circRNAs can not only predict tumor progression, but also serve as potential diagnostic biomarkers. Zheng et al demonstrated that hsa_circ_0015286 is significantly highly expressed in GC cells, tissues and plasma compared with normal controls and the AUC is 0.778, which is higher compared with the AUC of traditional tumor markers CA 19-9 and CEA (118). In addition, the AUC can be as high as 0.843 when the three indicators are combined. This study shows that exosomal hsa_circ_0015286 can be used in combination with traditional tumor markers CEA and CA 19-9 for high diagnostic efficacy, and it can also be diagnosed alone (118). Li et al examined the high expression of CDR1 antisense RNA (CDR1as) in the plasma and exosomes of patients with GC, and heat shock protein family E member 1 may be a key protein in the regulation of miRNA by CDR1as (119). These authors evaluated the AUC of tissue, plasma and exosomal derived CDR1as and revealed that tissue-derived CDR1as has the highest AUC of 0.782. The AUC of plasma CDR1as combined with conventional tumor markers is higher compared with that of tissue-derived CDR1as. This study suggests that CDR1as may be a potential diagnostic biomarker for GC (119). Tao et al showed that hsa_circ_0000419 is stably present in plasma exosomes and has low expression in GC tissues and cells. It regulates the development of GC by interacting with hsa-miR-589-3p and hsa-miR-141-5p. When the critical value of hsa_circ_0000419 is 4.90, the specificity and sensitivity of plasma hsa_circ_0000419 reaches 0.884 and 0.682, respectively (120).
Figure 4.
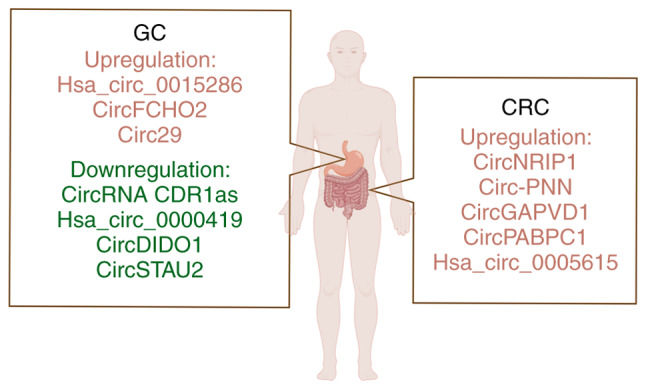
Exosomal circRNAs as new biomarkers and potential therapeutic targets of gastrointestinal cancer. By Figdraw (https://www.figdraw.com/#/). GC, gastric cancer; CRC, colorectal cancer; circRNA, circular RNA.
Table II.
Diagnostic and therapeutic significance of exosomal circRNAs in gastrointestinal cancer.
| Cancer type | CircRNA | CircBase ID | Expression | Source | Case no. | Potential function | AUC | Sensitivity (%), specificity (%) | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|
| GC | Hsa_circ_0015286 | Hsa_circ_0015286 | ↑ | Plasma- | 60 | Diagnosis and | 0.778 | 82.1, 65.7 | (118) |
| derived | prognosis | ||||||||
| EVs | |||||||||
| GC | CircRNA CDR1as | - | ↓ | Plasma- | 65 | Diagnosis and | 0.786 | - | (119 |
| derived | prognosis | ||||||||
| EVs | |||||||||
| GC | Hsa_circ_0000419 | Hsa_circ_0000419 | ↓ | Plasma- | 96 | Diagnosis and | 0.840 | 68.2, 88.4 | (120) |
| derived | prognosis | ||||||||
| EVs | |||||||||
| CRC | CircNRIP1 | Hsa_circ_0004771 | ↑ | Serum- | 170 | Diagnosis | 0.816 | 81.4, 74.3 | (121) |
| derived | |||||||||
| EVs | |||||||||
| CRC | Circ-PNN | Hsa_circ_0101802 | ↑ | Serum- | 88 | Diagnosis | 0.855 | 89.8, 73.9 | (122) |
| derived | |||||||||
| EVs | |||||||||
| CRC | CircGAPVD1 | Hsa_circ_0003270 | ↑ | Plasma- | 78 | Diagnosis | 0.7662 | 75.6, 71.7 | (123) |
| derived | |||||||||
| EVs | |||||||||
| GC | CircFCHO2 | - | ↑ | Serum- | - | Potential | - | - | (126) |
| derived | therapeutic | ||||||||
| EVs | target | ||||||||
| GC | Circ29 | Hsa_circ_0044366 | ↑ | Plasma- | - | Potential | - | - | (127) |
| derived | therapeutic | ||||||||
| EVs | target | ||||||||
| CRC | CircPABPC1 | Has-circ-0085159 | ↑ | Serum- | 60 | Potential | - | - | (128) |
| derived | therapeutic | ||||||||
| EVs | target | ||||||||
| GC | CircDIDO1 | Hsa_circ_0061137 | ↓ | Cell | 17 | Potential | - | - | (130) |
| superna | therapeutic | ||||||||
| tant- | target | ||||||||
| derived | |||||||||
| EVs | |||||||||
| GC | CircSTAU2 | Hsa_circ_0001811 | ↓ | Cell | 56 | Potential | - | - | (131) |
| superna | therapeutic | ||||||||
| tant- | target | ||||||||
| derived | |||||||||
| EVs | |||||||||
| CRC | Hsa_circ_0005615 | Hsa_circ_0005615 | ↑ | Serum- | 70 | Potential | - | - | (132) |
| derived | therapeutic | ||||||||
| EVs | target |
GC, gastric cancer; CRC, colorectal cancer; circRNA, circular RNA; AUC, area under ROC curve; EV, extracellular vesicle; ↑, upregulation; ↓, downregulation.
Furthermore, there are differences in the expression of exosomal circRNAs in CRC. Pan et al revealed that exosome-derived hsa-circ-0004771 is lowly expressed in CRC cells and tissues, but highly expressed in the serum of patients with CRC (121). The authors suggest that hsa-circ-000477 may bind to RNA-binding proteins and be transported to exosomes, which may be a mechanism for circRNA clearance. In different periods of CRC, the AUC of hsa-circ-000477 is >0.8, indicating that hsa-circ-000477 can be used as a novel potential diagnostic biomarker for CRC (121). According to Xie et al, circ-PNN is considerably overexpressed and strongly correlated with the prognosis of patients with CRC (122). The authors revealed that circ-PNN level has high performance in being able to differentiate patients with CRC from healthy controls; in addition, this study further investigated the possible clinical importance of serum exosomal circ-PNN. Circ-PNN might therefore be used as a diagnostic biomarker for CRC (122). Li et al revealed that circGAPVD1 is highly expressed in the plasma exosomes of CRC, with an AUC area of 0.7662 and diagnostic specificity and sensitivity of 71.79 and 75.64%, respectively (123). These results indicate that circGAPVD1 is expected to be a diagnostic marker for CRC (123). The aforementioned findings indicate that exosomal circRNAs are expected to be a novel biomarker for the prediction and diagnosis of gastrointestinal cancer.
5. Exosomal circRNAs as therapeutic targets of gastrointestinal cancer
Currently, the first-line treatment strategy for gastrointestinal cancer is chemotherapy, but cancer cells may become resistant to anticancer drugs or their targets. Finding novel treatment strategies at the molecular level is therefore crucial. Increasing evidence suggests that exosomal circRNAs are expected to be promising therapeutic targets for gastrointestinal cancer (124,125). Zhang et al revealed that circFCHO2 is highly expressed in serum exosomes of patients with GC (126). CircFCHO2 regulates the JAK1/STAT3 pathway by binding to miR-194-5p, which affects some malignant behaviors of GC cells; therefore, circFCHO2 may be a novel therapeutic target and diagnostic biomarker for GC (126). Li et al also demonstrated that exosomal-derived circ29 is substantially expressed in GC, which influences the GC formation by regulating the miR-29a/vascular endothelial growth factor axis, and is anticipated to emerge as a novel tumor marker and promising therapeutic target (127). In addition, Li et al demonstrated that exosome-derived circPABPC1 is highly expressed in CRC tissues and exist in the cytoplasm (128). CircPABPC1 promotes CRC progression by promoting the expression of important regulators of liver metastasis in CRC such as a disintegrin and metalloproteinase 19 and bone morphogenetic protein 4. Therefore, exosome-derived circPABPC1 is expected to be a novel biomarker and anti-metastatic therapeutic target for CRC (128). These circRNAs promote the malignant transformation of cancer cells, and inhibition of these exosomal circRNAs may help to suppress gastrointestinal cancers. In addition, it has been reported that exosome-mediated circRNA delivery with anti-tumor effects is considered to be an effective therapeutic method (109,129). For example, the results of Guo et al show that circDIDO1 inhibits GC progression by regulating the miR-1307-3p/suppressor of cytokine signaling 2 axis (130). Systemic administration of RGD-modified circDIDO1-loaded exosomes inhibits GC tumorigenicity and invasiveness in vitro and in vivo, suggesting that RGD-Exo-circDIDO1 can be used as a viable nanofat for GC treatment (130). Zhang et al demonstrated that exosome-delivered circSTAU2 can act as a tumor suppressor and inhibit GC progression through the miR-589/capping actin protein of muscle Z-line subunit α 1 axis, which indicates that circSTAU2 may be a potential therapeutic target for GC (131). Moreover, in CRC, it has been shown that circ_0005615 from serum exosomes of patients with CRC can modulate CRC malignant progression by controlling fos-like antigen 2 expression through sponging miR-873-5p, which reveals circ_0005615 as a new candidate target for CRC treatment (132). In summary, the treatment of gastrointestinal cancer mediated by exosomal circRNAs is still in its infancy, and the specific mechanism and safety of this strategy need to be further studied.
6. Challenges and perspectives
Exosomal circRNAs have a significant role in gastrointestinal cancers, as research has revealed in recent years (133,134). It has been demonstrated that circRNA has an impact on the proliferation, metastasis, stemness, angiogenesis, immune microenvironment and drug resistance of gastrointestinal tumors, and may develop into a useful therapeutic target for gastrointestinal malignancies as well as a potential biomarker (135). Although more studies have focused on the application of exosome-derived circRNAs in gastrointestinal tumors, there are still a number of problems to be solved.
First of all, existing studies lack technologies for rapid extraction and detection of exosomes and exosomal circRNAs. Currently, ultracentrifugation, density gradient centrifugation, ultrafiltration, immunoaffinity and polymer precipitation are the major methods used to isolate exosomes (136–138). Ultracentrifugation, the most commonly employed technique, is time-consuming and inconvenient and can lead to the destruction of exosomes. Exosomes obtained by ultracentrifugation have low purity and low yield due to the contamination of non-exosome components (136). So far, there is no clear method to directly extract circRNA, but total RNA can be digested with RNase R to enrich circRNA. Second, the secretion mechanism of exosomes-derived circRNAs remains unclear. Future research should concentrate on how exosomes discharge circRNAs to act on the desired cells. In addition, the limited number of circRNAs with clear mechanisms of action in gastrointestinal cancer requires further research and exploration. Finally, there is still a lack of a large number of prospective studies on exosome-derived circRNA as a diagnostic marker and targeted therapy molecule for gastrointestinal tumors. It is important to further assess the sensitivity, specificity and diagnostic impact of exosomal circRNAs paired with current tumor markers as a diagnostic marker. Exosomal circRNAs, which is a targeted therapeutic molecule, encounters more difficulties with regard to its efficacy, safety, potential side effects, delivery method, biodistribution and effective dose.
7. Conclusions
In conclusion, exosomal circRNAs have promise as novel diagnostic biomarkers and therapeutic targets and play a critical role in the development of gastrointestinal malignancies. It will be beneficial for its clinical use if the study of exosomal circRNAs and gastrointestinal cancers becomes more in-depth and cutting-edge technologies, including exosomal circRNA separation and detection, are developed. The present study hypothesizes that diagnostic and therapeutic strategies based on exosomal circRNAs can be effectively applied to clinical practice in the future.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. 81602883), the Project of Social Development in Zhenjiang (grant no. SH2021045), the Technology Development Foundation of Jiangsu University (grant no. 20220516), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant no. KYCX23_3765) and the Student Innovation Training Program of Jiangsu University (grant nos. 202310299410X and 202310299476X).
Availability of data and materials
Not applicable.
Authors' contributions
ZL, JJ and HQ designed research and wrote the paper. YX, XuZ, XiZ, JS, XZ and ZG have made contributions to analysis and interpretation of data. XuZ, XiZ, JS, XZ and ZG have made contributions to the production of tables and figures. YX, JH and XiZ contributed to the writing and revisions. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
References
- 1.Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159:335–349.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, Sung B, Aggarwal BB. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res. 2008;25:2097–2116. doi: 10.1007/s11095-008-9690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kashyap S, Pal S, Chandan G, Saini V, Chakrabarti S, Saini NK, Mittal A, Thakur VK, Saini AK, Saini RV. Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Semin Cancer Biol. 2022;86((Pt 3)):643–651. doi: 10.1016/j.semcancer.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Wang D, Zhang C, Liu H, Hao M, Kan S, Liu D, Liu W. The applications of gold nanoparticles in the diagnosis and treatment of gastrointestinal cancer. Front Oncol. 2022;11:819329. doi: 10.3389/fonc.2021.819329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang DM, Chan KKW, Jang RW, Booth C, Liu G, Amir E, Mason R, Everest L, Elimova E. Anticancer drugs approved by the Food and Drug Administration for gastrointestinal malignancies: Clinical benefit and price considerations. Cancer Med. 2019;8:1584–1593. doi: 10.1002/cam4.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh D, Dheer D, Samykutty A, Shankar R. Antibody drug conjugates in gastrointestinal cancer: From lab to clinical development. J Control Release. 2021;340:1–34. doi: 10.1016/j.jconrel.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M, Hu S, Liu L, Dang P, Liu Y, Sun Z, Qiao B, Wang C. Engineered exosomes from different sources for cancer-targeted therapy. Signal Transduct Target Ther. 2023;8:124. doi: 10.1038/s41392-023-01382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21:799–820. doi: 10.1038/s41573-022-00520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi MAMA, Almazyadi HAM, Kallmeyer K, Dandara C, Pepper MS, et al. The role of tumor microenvironment in chemoresistance: To survive, keep your enemies closer. Int J Mol Sci. 2017;18:1586. doi: 10.3390/ijms18071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20:840. doi: 10.3390/ijms20040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. doi: 10.1080/2162402X.2015.1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung BH, von Lersner A, Guerrero J, Krystofiak ES, Inman D, Pelletier R, Zijlstra A, Ponik SM, Weaver AM. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat Commun. 2020;11:2092. doi: 10.1038/s41467-020-15747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pluchino S, Smith JA. Explicating exosomes: Reclassifying the rising stars of intercellular communication. Cell. 2019;177:225–227. doi: 10.1016/j.cell.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Huang W, Li M, Zheng A. Exosome-Based carrier for RNA delivery: Progress and challenges. Pharmaceutics. 2023;15:598. doi: 10.3390/pharmaceutics15020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng X, Xiao W, Sun J, Li W, Yuan H, Yu T, Zhang X, Dong W. CircPTK2/PABPC1/SETDB1 axis promotes EMT-mediated tumor metastasis and gemcitabine resistance in bladder cancer. Cancer Lett. 2023;554:216023. doi: 10.1016/j.canlet.2022.216023. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Zhang N, Chai J, Wang T, Ma C, Han L, Yang M. CircPDIA4 induces gastric cancer progression by promoting ERK1/2 activation and enhancing biogenesis of oncogenic circRNAs. Cancer Res. 2023;83:538–552. doi: 10.1158/0008-5472.CAN-22-1923. [DOI] [PubMed] [Google Scholar]
- 18.Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, Li X, Lin C. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20:26. doi: 10.1186/s12943-021-01318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Lin YL, Shao JK, Wu XJ, Li X, Yao H, Shi FL, Li LS, Zhang WG, Chang ZY, et al. Plasma exosomal hsa_circ_0079439 as a novel biomarker for early detection of gastric cancer. World J Gastroenterol. 2023;29:3482–3496. doi: 10.3748/wjg.v29.i22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang XJ, Wang Y, Wang HT, Liang ZF, Ji C, Li XX, Zhang LL, Ji RB, Xu WR, Jin JH, Qian H. Exosomal hsa_circ_000200 as a potential biomarker and metastasis enhancer of gastric cancer via miR-4659a/b-3p/HBEGF axis. Cancer Cell Int. 2023;23:151. doi: 10.1186/s12935-023-02976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing L, Xia M, Jiao X, Fan L. Hsa_circ_0004831 serves as a blood-based prognostic biomarker for colorectal cancer and its potentially circRNA-miRNA-mRNA regulatory network construction. Cancer Cell Int. 2020;20:557. doi: 10.1186/s12935-020-01651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocucci E, Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Kok VC, Yu CC. Cancer-Derived Exosomes: Their role in cancer biology and biomarker development. Int J Nanomedicine. 2020;15:8019–8036. doi: 10.2147/IJN.S272378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, Zhang W, Pu F, Shao Z. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J Control Release. 2022;343:107–117. doi: 10.1016/j.jconrel.2022.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 28.Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Zhang H, Gu J, Zhang J, Shi H, Qian H, Wang D, Xu W, Pan J, Santos HA. Engineered extracellular vesicles for cancer therapy. Adv Mater. 2021;33:e2005709. doi: 10.1002/adma.202005709. [DOI] [PubMed] [Google Scholar]
- 31.Tallon C, Hollinger KR, Pal A, Bell BJ, Rais R, Tsukamoto T, Witwer KW, Haughey NJ, Slusher BS. Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases. Drug Discov Today. 2021;26:1656–1668. doi: 10.1016/j.drudis.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60:9–18. doi: 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson G. Isolation of lipid rafts (Detergent-Resistant Microdomains) and comparison to extracellular vesicles (Exosomes) Methods Mol Biol. 2021;2187:99–112. doi: 10.1007/978-1-0716-0814-2_6. [DOI] [PubMed] [Google Scholar]
- 35.Parton RG, McMahon KA, Wu Y. Caveolae: Formation, dynamics, and function. Curr Opin Cell Biol. 2020;65:8–16. doi: 10.1016/j.ceb.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Kwiatkowska K, Matveichuk OV, Fronk J, Ciesielska A. Flotillins: At the Intersection of Protein S-Palmitoylation and lipid-mediated signaling. Int J Mol Sci. 2020;21:2283. doi: 10.3390/ijms21072283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikonen E, Zhou X. Cholesterol transport between cellular membranes: A balancing act between interconnected lipid fluxes. Dev Cell. 2021;56:1430–1436. doi: 10.1016/j.devcel.2021.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Kummer D, Steinbacher T, Schwietzer MF, Thölmann S, Ebnet K. Tetraspanins: integrating cell surface receptors to functional microdomains in homeostasis and disease. Med Microbiol Immunol. 2020;209:397–405. doi: 10.1007/s00430-020-00673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YJ, Shin KJ, Jang HJ, Ryu JS, Lee CY, Yoon JH, Seo JK, Park S, Lee S, Je AR, et al. GPR143 controls ESCRT-dependent exosome biogenesis and promotes cancer metastasis. Dev Cell. 2023;58:320–334.e8. doi: 10.1016/j.devcel.2023.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Qi R, Bai Y, Li K, Liu N, Xu Y, Dal E, Wang Y, Lin R, Wang H, Liu Z, et al. Cancer-associated fibroblasts suppress ferroptosis and induce gemcitabine resistance in pancreatic cancer cells by secreting exosome-derived ACSL4-targeting miRNAs. Drug Resist Updat. 2023;68:100960. doi: 10.1016/j.drup.2023.100960. [DOI] [PubMed] [Google Scholar]
- 41.He Z, Wang J, Zhu C, Xu J, Chen P, Jiang X, Chen Y, Jiang J, Sun C. Exosome-derived FGD5-AS1 promotes tumor-associated macrophage M2 polarization-mediated pancreatic cancer cell proliferation and metastasis. Cancer Lett. 2022;548:215751. doi: 10.1016/j.canlet.2022.215751. [DOI] [PubMed] [Google Scholar]
- 42.Xu Z, Zeng S, Gong Z, Yan Y. Exosome-based immunotherapy: A promising approach for cancer treatment. Mol Cancer. 2020;19:160. doi: 10.1186/s12943-020-01278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, Chen X, Chen Y, Xu C, Hu Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054–1070. doi: 10.1016/j.ymthe.2022.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomes C, Keller S, Altevogt P, Costa J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci Lett. 2007;428:43–46. doi: 10.1016/j.neulet.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 47.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Z, Cai Y, Liu W, Kang F, He Q, Hong Q, Zhang W, Li J, Yan Y, Peng J. Downregulated exosome-associated gene FGF9 as a novel diagnostic and prognostic target for ovarian cancer and its underlying roles in immune regulation. Aging (Albany NY) 2022;14:1822–1835. doi: 10.18632/aging.203905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Cao Y, Sun M, Feng H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 2021;35:e21916. doi: 10.1096/fj.202100294RR. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vea A, Llorente-Cortes V, de Gonzalo-Calvo D. Circular RNAs in Blood. Adv Exp Med Biol. 2018;1087:119–130. doi: 10.1007/978-981-13-1426-1_10. [DOI] [PubMed] [Google Scholar]
- 52.Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, Guo J. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11–20. doi: 10.2217/bmm-2017-0114. [DOI] [PubMed] [Google Scholar]
- 53.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 54.Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018;7:3101–3109. doi: 10.1002/cam4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng D, Luo L, Zhang X, Wei C, Zhang Z, Han L. CircRNA: An emerging star in the progression of glioma. Biomed Pharmacother. 2022;151:113150. doi: 10.1016/j.biopha.2022.113150. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Liu F, Feng Y, Xu X, Wang Y, Zhu S, Dong J, Zhao S, Xu B, Feng N. CircRNA circ_0006156 inhibits the metastasis of prostate cancer by blocking the ubiquitination of S100A9. Cancer Gene Ther. 2022;29:1731–1741. doi: 10.1038/s41417-022-00492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang D, Ni N, Wang Y, Tang Z, Gao H, Ju Y, Sun N, He X, Gu P, Fan X. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin α5 expression. Cell Death Differ. 2021;28:283–302. doi: 10.1038/s41418-020-0600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, Liu H, Jiang J, Yang Y, Wang W, Jia Z. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging (Albany NY) 2021;13:18978–18992. doi: 10.18632/aging.203347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang G, Liang M, Liu H, Huang J, Li P, Wang C, Zhang Y, Lin Y, Jiang X. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020;11:1065. doi: 10.1038/s41419-020-03276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, Kong X, Bu J, Liu M, Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, Huang N, Yang X, Xiao F, Liu D, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131. doi: 10.1186/s12943-019-1083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Begum S, Yiu A, Stebbing J, Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37:4055–4057. doi: 10.1038/s41388-018-0230-3. [DOI] [PubMed] [Google Scholar]
- 66.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 Is a Circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu H, Zhang Z, Yuan X, Song H, Li P. The role of circular RNA hsa_circ_0001789 as a diagnostic biomarker in gastric carcinoma. Scand J Gastroenterol. 2023;58:248–253. doi: 10.1080/00365521.2022.2122865. [DOI] [PubMed] [Google Scholar]
- 68.Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, Pourhanifeh MH, Savardashtaki A, Mirzaei H. Circular RNAs in cancer: New insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12:84. doi: 10.1186/s13048-019-0558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He Y, Zheng L, Yuan M, Fan J, Rong L, Zhan T, Zhang J. Exosomal circPRRX1 functions as a ceRNA for miR-596 to promote the proliferation, migration, invasion, and reduce radiation sensitivity of gastric cancer cells via the upregulation of NF-κB activating protein. Anticancer Drugs. 2022;33:1114–1125. doi: 10.1097/CAD.0000000000001358. [DOI] [PubMed] [Google Scholar]
- 72.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K, et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211–8226. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi H, Lee DS. Illuminating the physiology of extracellular vesicles. Stem Cell Res Ther. 2016;7:55. doi: 10.1186/s13287-016-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen C, Yu H, Han F, Lai X, Ye K, Lei S, Mai M, Lai M, Zhang H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21:46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan G, Ding W, Sun B, Zhu L, Gao Y, Chen L. Upregulated circRNA_102231 promotes gastric cancer progression and its clinical significance. Bioengineered. 2021;12:4936–4945. doi: 10.1080/21655979.2021.1960769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E, Gong C. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21:933. doi: 10.1186/s12885-021-08669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vakhshiteh F, Hassani S, Momenifar N, Pakdaman F. Exosomal circRNAs: New players in colorectal cancer. Cancer Cell Int. 2021;21:483. doi: 10.1186/s12935-021-02112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, Yang J, Chen M, Dang Y, Zhang G. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis. 2022;13:56. doi: 10.1038/s41419-021-04364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu L, Xie J, Liu X, Yu Y, Wang S. Plasma Exosomal CircNEK9 accelerates the progression of gastric cancer via miR-409-3p/MAP7 axis. Dig Dis Sci. 2021;66:4274–4289. doi: 10.1007/s10620-020-06816-z. [DOI] [PubMed] [Google Scholar]
- 84.Fan L, Li W, Jiang H. Circ_0000395 Promoted CRC Progression via Elevating MYH9 Expression by Sequestering miR-432-5p. Biochem Genet. 2023;61:116–137. doi: 10.1007/s10528-022-10245-0. [DOI] [PubMed] [Google Scholar]
- 85.Gao L, Tang X, He Q, Sun G, Wang C, Qu H. Exosome-transmitted circCOG2 promotes colorectal cancer progression via miR-1305/TGF-β2/SMAD3 pathway. Cell Death Discov. 2021;7:281. doi: 10.1038/s41420-021-00680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 87.Hui C, Tian L, He X. Circular RNA circNHSL1 contributes to gastric cancer progression through the miR-149-5p/YWHAZ axis. Cancer Manag Res. 2020;12:7117–7130. doi: 10.2147/CMAR.S253152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen X, Kong S, Ma S, Shen L, Zheng M, Qin S, Qi J, Wang Q, Cui X, Ju S. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene. 2022;41:4724–4735. doi: 10.1038/s41388-022-02449-w. [DOI] [PubMed] [Google Scholar]
- 89.Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, Feng ZH, Luo J, Jin XH, Duan JL, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen C, Liu Y, Liu L, Si C, Xu Y, Wu X, Wang C, Sun Z, Kang Q. Exosomal circTUBGCP4 promotes vascular endothelial cell tipping and colorectal cancer metastasis by activating Akt signaling pathway. J Exp Clin Cancer Res. 2023;42:46. doi: 10.1186/s13046-023-02619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu Q, Zhang Y, Tian Y, Peng A, Cui X, Ding B, Yang L, Liu Y, Ju Y, Gao C. Exosomal Circ_FMN2 derived from the serum of colorectal cancer patients promotes cancer progression by miR-338-3p/MSI1 Axis. Appl Biochem Biotechnol. 2023 Mar 30; doi: 10.1007/s12010-023-04722-4. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 92.Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li YF, Pei FL, Cao MZ. CircRNA_101951 promotes migration and invasion of colorectal cancer cells by regulating the KIF3A-mediated EMT pathway. Exp Ther Med. 2020;19:3355–3361. doi: 10.3892/etm.2020.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22:2297–2303. doi: 10.26355/eurrev_201804_14818. [DOI] [PubMed] [Google Scholar]
- 95.Liang ZF, Zhang Y, Guo W, Chen B, Fang S, Qian H. Gastric cancer stem cell-derived exosomes promoted tobacco smoke-triggered development of gastric cancer by inducing the expression of circ670. Med Oncol. 2022;40:24. doi: 10.1007/s12032-022-01906-6. [DOI] [PubMed] [Google Scholar]
- 96.Miao Z, Zhao X, Liu X. Exosomal circCOL1A2 from cancer cells accelerates colorectal cancer progression via regulating miR-665/LASP1 signal axis. Eur J Pharmacol. 2023;950:175722. doi: 10.1016/j.ejphar.2023.175722. [DOI] [PubMed] [Google Scholar]
- 97.Zhao H, Chen S, Fu Q. Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J Cell Biochem. 2020;121:3286–3297. doi: 10.1002/jcb.29600. [DOI] [PubMed] [Google Scholar]
- 98.Liu C, Yang J, Zhu F, Zhao Z, Gao L. Exosomal circ_0001190 regulates the progression of gastric cancer via miR-586/SOSTDC1 Axis. Biochem Genet. 2022;60:1895–1913. doi: 10.1007/s10528-021-10180-6. [DOI] [PubMed] [Google Scholar]
- 99.Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020;14:2960–2984. doi: 10.1002/1878-0261.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song J, Xu X, He S, Wang N, Bai Y, Li B, Zhang S. Exosomal hsa_circ_0017252 attenuates the development of gastric cancer via inhibiting macrophage M2 polarization. Hum Cell. 2022;35:1499–1511. doi: 10.1007/s13577-022-00739-9. [DOI] [PubMed] [Google Scholar]
- 101.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng Y, Li Z, Wang Y, Chen W, Lin Y, Guo J, Ye G. CircRNA: A new class of targets for gastric cancer drug resistance therapy. Pathol Oncol Res. 2023;29:1611033. doi: 10.3389/pore.2023.1611033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu D, Chang Z, Liu X, Chen P, Zhang H, Qin Y. Macrophage-derived exosomes regulate gastric cancer cell oxaliplatin resistance by wrapping circ 0008253. Cell Cycle. 2023;22:705–717. doi: 10.1080/15384101.2022.2146839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Y, Liu H, Zou J, Cao G, Li Y, Xing C, Wu J. Exosomal circ_0091741 promotes gastric cancer cell autophagy and chemoresistance via the miR-330-3p/TRIM14/Dvl2/Wnt/β-catenin axis. Hum Cell. 2023;36:258–275. doi: 10.1007/s13577-022-00790-6. [DOI] [PubMed] [Google Scholar]
- 105.Yang G, Tan J, Guo J, Wu Z, Zhan Q. Exosome-mediated transfer of circ_0063526 enhances cisplatin resistance in gastric cancer cells via regulating miR-449a/SHMT2 axis. Anticancer Drugs. 2022;33:1047–1057. doi: 10.1097/CAD.0000000000001386. [DOI] [PubMed] [Google Scholar]
- 106.Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis Via miR-30a-5p/YAP1 axis in gastric cancer cells. Cancer Biother Radiopharm. 2021;36:347–359. doi: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- 107.Pan Z, Zheng J, Zhang J, Lin J, Lai J, Lyu Z, Feng H, Wang J, Wu D, Li Y. A novel protein encoded by exosomal CircATG4B induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv Sci (Weinh) 2022;9:e2204513. doi: 10.1002/advs.202204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y, Tan X, Lu Y. Exosomal transfer of circ_0006174 contributes to the chemoresistance of doxorubicin in colorectal cancer by depending on the miR-1205/CCND2 axis. J Physiol Biochem. 2022;78:39–50. doi: 10.1007/s13105-021-00831-y. [DOI] [PubMed] [Google Scholar]
- 109.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao K, Cheng X, Ye Z, Li Y, Peng W, Wu Y, Xing C. Exosome-Mediated Transfer of circ_0000338 Enhances 5-Fluorouracil resistance in colorectal cancer through regulating MicroRNA 217 (miR-217) and miR-485-3p. Mol Cell Biol. 2021;41:e00517–20. doi: 10.1128/MCB.00517-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2019;16:282–295. doi: 10.1038/s41575-019-0115-0. [DOI] [PubMed] [Google Scholar]
- 112.Xu R, Greening DW, Zhu HJ, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: Toward clinical application. J Clin Invest. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 114.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 115.Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, Xu X, Liang Q, Christiani DC, Wang M, et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi: 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325. doi: 10.1186/s13046-018-1006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zheng P, Gao H, Xie X, Lu P. Plasma Exosomal hsa_circ_0015286 as a potential diagnostic and prognostic biomarker for gastric cancer. Pathol Oncol Res. 2022;28:1610446. doi: 10.3389/pore.2022.1610446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li R, Tian X, Jiang J, Qian H, Shen H, Xu W. CircRNA CDR1as: a novel diagnostic and prognostic biomarker for gastric cancer. Biomarkers. 2023;28:448–457. doi: 10.1080/1354750X.2023.2206984. [DOI] [PubMed] [Google Scholar]
- 120.Tao X, Shao Y, Lu R, Ye Q, Xiao B, Ye G, Guo J. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract. 2020;216:152763. doi: 10.1016/j.prp.2019.152763. [DOI] [PubMed] [Google Scholar]
- 121.Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, et al. Identification of Serum Exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, Zhao Y, Duan W, Chen Y, Wang Y, et al. RNA-Seq profiling of serum exosomal circular RNAs Reveals Circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol. 2020;10:982. doi: 10.3389/fonc.2020.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li T, Zhou T, Wu J, Lv H, Zhou H, Du M, Zhang X, Wu N, Gong S, Ren Z, et al. Plasma exosome-derived circGAPVD1 as a potential diagnostic marker for colorectal cancer. Transl Oncol. 2023;31:101652. doi: 10.1016/j.tranon.2023.101652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang H, Zeng X, Zheng Y, Wang Y, Zhou Y. Exosomal circRNA in digestive system tumors: the main player or coadjuvants? Front Oncol. 2021;11:614462. doi: 10.3389/fonc.2021.614462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu L, Fang S, Zhang Y, Jin L, Xu W, Liang Z. Exosomes and Exosomal circRNAs: The rising stars in the progression, diagnosis and prognosis of gastric cancer. Cancer Manag Res. 2021;13:8121–8129. doi: 10.2147/CMAR.S331221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Z, Sun C, Zheng Y, Gong Y. circFCHO2 promotes gastric cancer progression by activating the JAK1/STAT3 pathway via sponging miR-194-5p. Cell Cycle. 2022;21:2145–2164. doi: 10.1080/15384101.2022.2087280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li S, Li J, Zhang H, Zhang Y, Wang X, Yang H, Zhou Z, Hao X, Ying G, Ba Y. Gastric cancer derived exosomes mediate the delivery of circRNA to promote angiogenesis by targeting miR-29a/VEGF axis in endothelial cells. Biochem Biophys Res Commun. 2021;560:37–44. doi: 10.1016/j.bbrc.2021.04.099. [DOI] [PubMed] [Google Scholar]
- 128.Li Y, Hu J, Wang M, Yuan Y, Zhou F, Zhao H, Qiu T, Liang L. Exosomal circPABPC1 promotes colorectal cancer liver metastases by regulating HMGA2 in the nucleus and BMP4/ADAM19 in the cytoplasm. Cell Death Discov. 2022;8:335. doi: 10.1038/s41420-022-01124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yi Q, Yue J, Liu Y, Shi H, Sun W, Feng J, Sun W. Recent advances of exosomal circRNAs in cancer and their potential clinical applications. J Transl Med. 2023;21:516. doi: 10.1186/s12967-023-04348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guo Z, Zhang Y, Xu W, Zhang X, Jiang J. Engineered exosome-mediated delivery of circDIDO1 inhibits gastric cancer progression via regulation of MiR-1307-3p/SOCS2 Axis. J Transl Med. 2022;20:326. doi: 10.1186/s12967-022-03527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang C, Wei G, Zhu X, Chen X, Ma X, Hu P, Liu W, Yang W, Ruan T, Zhang W, et al. Exosome-Delivered circSTAU2 inhibits the progression of gastric cancer by targeting the miR-589/CAPZA1 Axis. Int J Nanomedicine. 2023;18:127–142. doi: 10.2147/IJN.S391872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu L, Zhang F, Wang Y. Circ_0005615 regulates the progression of colorectal cancer through the miR-873-5p/FOSL2 signaling pathway. Biochem Genet. 2023;61:2020–2041. doi: 10.1007/s10528-023-10355-3. [DOI] [PubMed] [Google Scholar]
- 133.Meng X, Yang D, Zhang B, Zhao Y, Zheng Z, Zhang T. Regulatory mechanisms and clinical applications of tumor-driven exosomal circRNAs in cancers. Int J Med Sci. 2023;20:818–835. doi: 10.7150/ijms.82419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lei T, Zhang Y, Wang X, Liu W, Feng W, Song W. Integrated analysis of the functions and clinical implications of exosome circRNAs in colorectal cancer. Front Immunol. 2022;13:919014. doi: 10.3389/fimmu.2022.919014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang F, Jiang J, Qian H, Yan Y, Xu W. Exosomal circRNA: Emerging insights into cancer progression and clinical application potential. J Hematol Oncol. 2023;16:67. doi: 10.1002/hon.3163_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lin S, Yu Z, Chen D, Wang Z, Miao J, Li Q, Zhang D, Song J, Cui D. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16:e1903916. doi: 10.1002/smll.201903916. [DOI] [PubMed] [Google Scholar]
- 137.Chen L, Wang L, Zhu L, Xu Z, Liu Y, Li Z, Zhou J, Luo F. Exosomes as drug carriers in anti-cancer therapy. Front Cell Dev Biol. 2022;10:728616. doi: 10.3389/fcell.2022.728616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ding L, Yang X, Gao Z, Effah CY, Zhang X, Wu Y, Qu L. A holistic review of the state-of-the-art microfluidics for exosome separation: An overview of the current status, existing obstacles, and future outlook. Small. 2021;17:e2007174. doi: 10.1002/smll.202007174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.