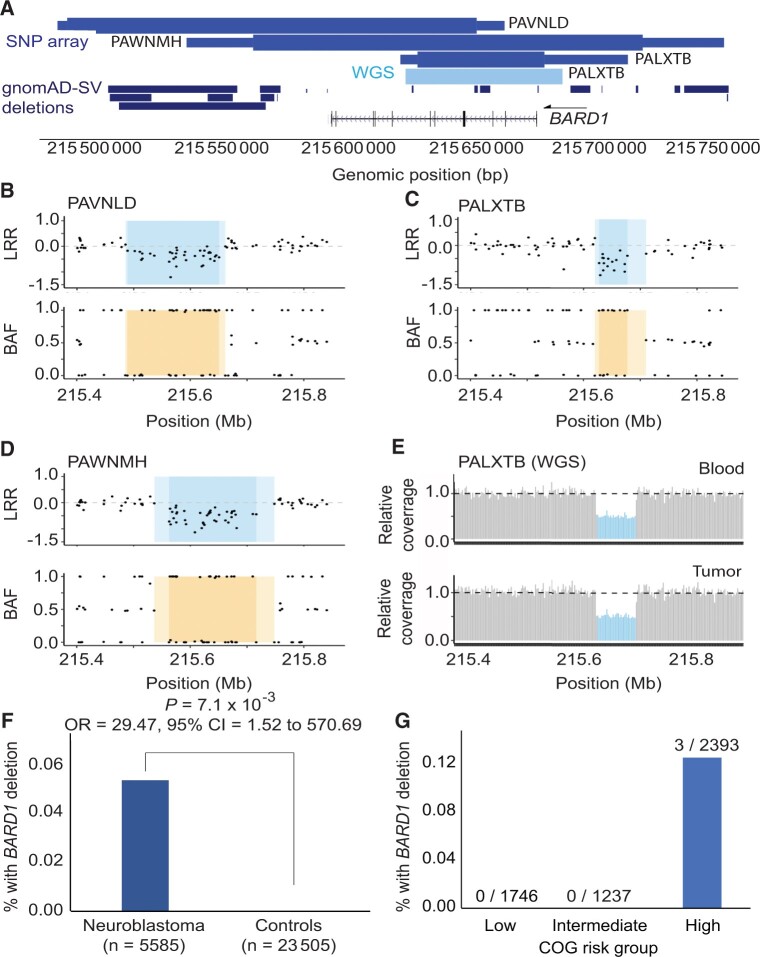
Figure 3.
Rare germline copy number variants disrupting BARD1 in neuroblastoma patients. A) BARD1 deletions were identified in 3 of 5585 neuroblastoma patients through copy number analysis of a large germline single nucleotide polymorphism (SNP) array dataset (medium blue, top track). No deletions were observed in 23 505 array-matched cancer-free control participants. The thick and thin bars represent minimum and maximum deletion coordinates, respectively. One deletion was validated and fine-mapped by whole genome sequencing (WGS; light blue, middle track). No BARD1 protein coding deletions were observed in 10 847 individuals in the gnomAD v2.1 structural variant dataset (dark blue, bottom track). B-D) The 3 array-based copy number variant calls are shown in log R ratio (LRR) and B allele frequency (BAF) plots. Darker shading indicates the minimum deleted region, whereas lighter shading indicates the maximum region. E) WGS validation for patient PALXTB is shown as relative sequencing coverage for matched blood and tumor samples. F) Rare BARD1 deletion copy number variants are enriched in neuroblastoma compared to cancer-free participants. G) Deletions disrupting BARD1 were observed exclusively in patients diagnosed with high-risk subset of neuroblastoma. gnomAD = Genome Aggregation Database.