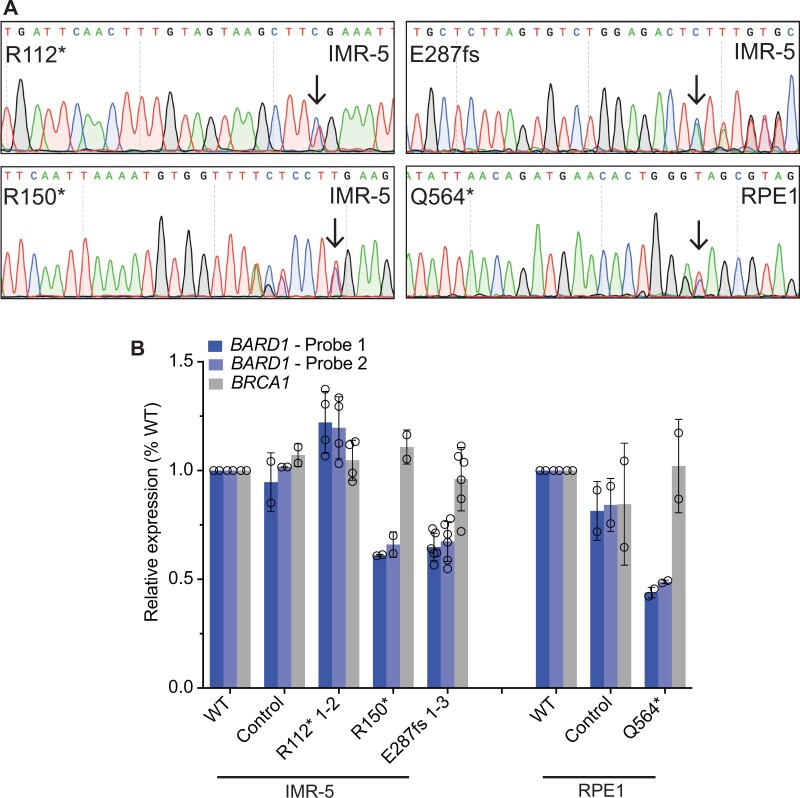
Figure 2.
Neuroblastoma cells heterozygous for disease-associated BARD1 loss-of-function variants (BARD1+/mut) have reduced BARD1 expression. A) Representative chromatograms from IMR-5 and RPE1 BARD1+/mut isogenic cell lines. Black arrows indicate CRISPR-introduced BARD1 heterozygous variants. Other variants reflect synonymous protospacer adjacent motif (PAM) changes (R150*, Q564*) or frameshift-induced nucleotide alterations (E287fs). B) BARD1 and BRCA1 expression in IMR-5 BARD1+/mut (left) and RPE1 BARD1+/mut (right) cells and nontargeted clonal control cells. BARD1 expression measured via 2 unique TaqMan probes. Data in panel B is represented as means (SD) of 2 biological replicates of each unique cell line, including multiple cell lines with identical BARD1 variants (n = 2 IMR-5 BARD1+/R112*; n = 1 for IMR-5 BARD1+/R150*; and n = 3 for IMR-5 BARD1+/E287fs cell lines). WT = wild-type.