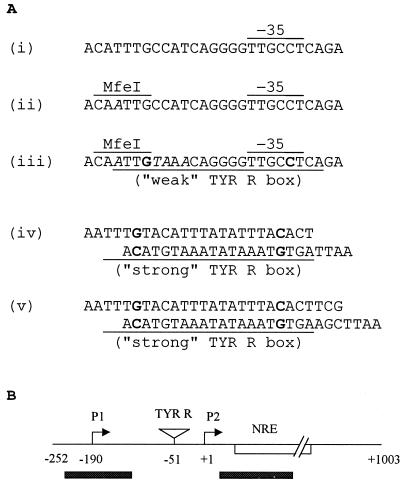
FIG. 1.
Introduction of the TYR R box(es) near proU P2. (A) The nucleotide sequence upstream of the −35 region of the wild-type proU P2 promoter (i) and those following sequential site-directed mutagenesis to create first an MfeI site (at position −51 relative to the start site of P2 transcription) (ii) and then a weak TYR R box (iii) are shown. The MfeI and the −35 hexamer sequences are indicated, and the mutated base residues are in italics. Also shown are the pairs of annealed oligonucleotide sequences (iv and v) that were used to generate the double-stranded TYR R strong box sequences (identical to that in tyrP) flanked with 5′-AATT overhangs for construction of the repression tester and activation tester variants, following insertion into the MfeI sites shown in sequences iii and ii, respectively. The 22-bp TYR R box sequences (whose consensus is the palindrome 5′-N2TGTAAAN6TTTACAN2-3′, in which the residues shown in bold are invariant) are underlined, and the invariant residues are in bold. (B) Schematic depiction of the position of insertion of the TYR R strong box sequences in the two proU variants, relative to P1, P2, and the NRE. For nucleotide numbering, the start site of P2 transcription has been taken to be +1. Shaded bars indicate the regions of DNA curvature to which H-NS exhibits preferential binding.