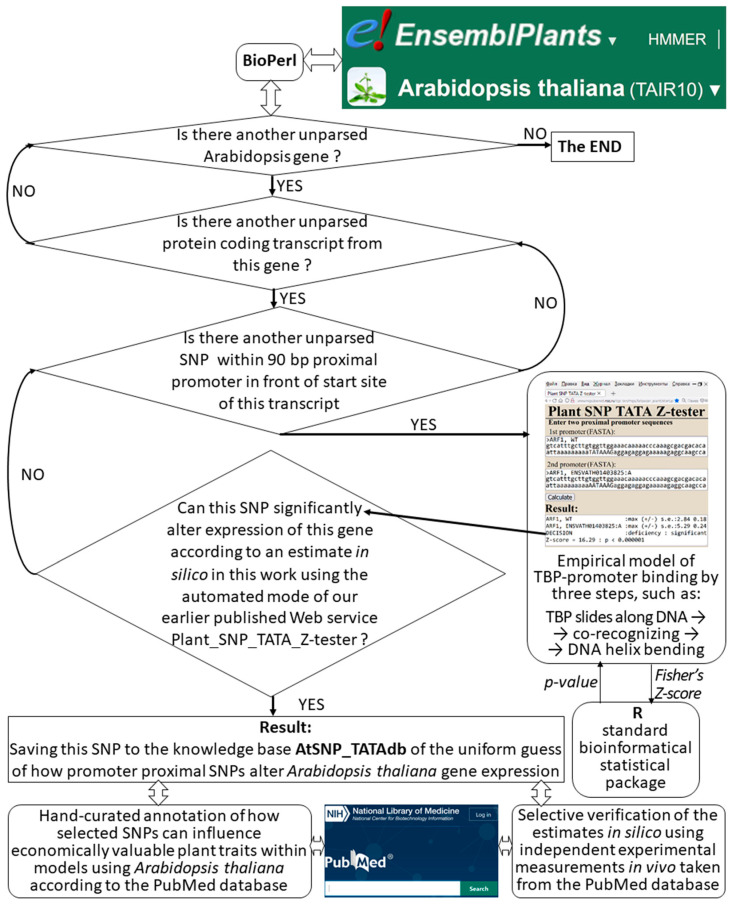
Figure 1.
Flowchart of AtSNP_TATAdb knowledge base development by processing genome-wide information from the Ensembl Plant database [31] by first using Plant_SNP_TATA_Z-tester [24] and, after that, selectively annotating some of the SNPs by a search in the PubMed database [39] using information about where, when and under what conditions changes in expression of these genes or their homolog genes were observed in agricultural studies. Finally, we selectively verified estimates in silico using independent experimental data in vivo taken from the PubMed database [39].