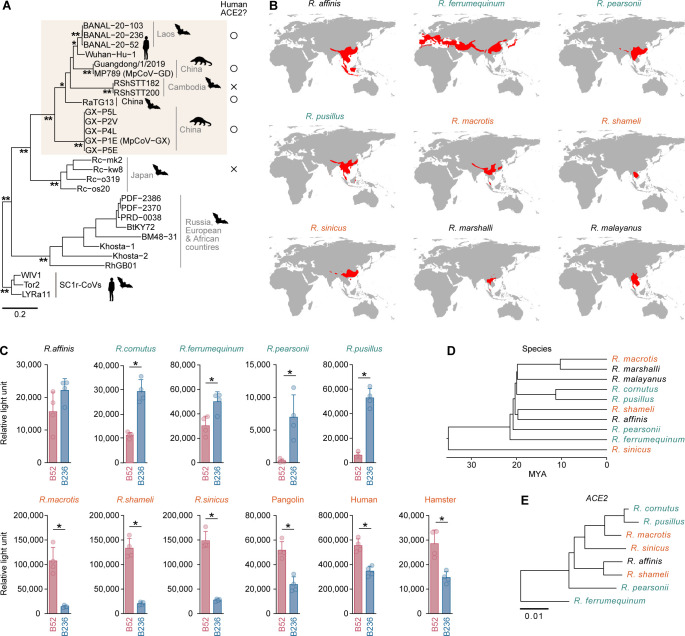
FIG 1.
Different ACE2 tropism of the two Laotian SC2r-CoVs. (A) Maximum likelihood tree of SC2r-CoVs and SARS-CoV-2 (strain Wuhan-Hu-1) based on their nucleotide sequences corresponding to RBD in S. SARS-CoV-1 (strain Tor2) and two SC1r-CoVs (WIV1 and LYRa11) are included as an outgroup. *, >0.8 bootstrap value; **, >0.9 bootstrap value. The scale bar indicates genetic distance. The usability of human ACE2 for SC2r-CoV infection is indicated with ○ (yes) or × (no), respectively, and the clade of SC2r-CoVs that can use human ACE2 is shaded in brown. (B) Geological distributions of Rhinolophus bat species. The habitat information originates from the IUCN Red List of Threatened Species website (https://www.iucnredlist.org/). Note that habitat information for R. cornutus is not available. Also, the habitat information for R. marshalli (B236 was isolated) and R. malayanus (B52 was isolated) is included. (C) Pseudovirus assay. HIV-1-based reporter viruses pseudotyped with the S proteins of B52 or B236 were prepared. The pseudoviruses were inoculated into a series of HOS-TMPRSS2 cells stably expressing Rhinolophus bat ACE2 cells at 1 ng HIV-1 p24 antigen. The infectivity (relative light unit) in each target cell is shown. The host species in which ACE2 is preferred by B52 or B236 are indicated in green and orange, respectively. Data are expressed as the mean with SD. Assays were performed in quadruplicate. Statistically significant differences (*P < 0.05) between B52 and B236 were determined by a two-sided Student’s t-test. (D and E) Phylogenetic relationship of Rhinolophus bat species. (D) Time-calibrated species tree for Rhinolophus bat species generated by TimeTree5 (16). MYA, million years ago. (E) Maximum likelihood tree of Rhinolophus bat ACE2 sequences. The scale bar indicates genetic distance.