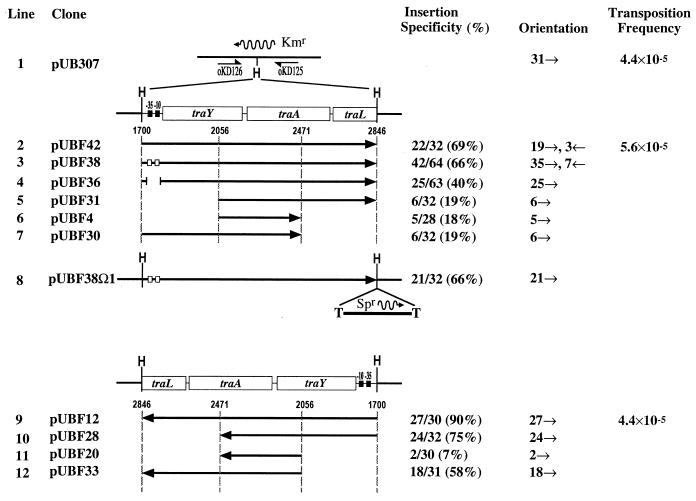
FIG. 5.
IS903 insertion specificity and orientation preference in region I from pOX38 when cloned into pUB307. A simple map of the Kmr gene in pUB307 is indicated in line 1. The direction of transcription of the Kmr gene is designated by the wavy arrow. Oligonucleotides oKD125 and oKD126 were used in a PCR analysis to identify IS903 insertions into the region. To determine the orientation of those insertions by PCR, oKD125 and oKD126 were used in combination with the transposon-specific primers oKD78 and oKD79 (Fig. 1). Region I and derivatives of it were cloned into the unique HindIII (H) site of pUB307 in both orientations (lines 2 to 12). The DNA present in each plasmid is shown by the solid arrow. The tra genes and their coordinates with respect to the BglII site in the pOX38 transfer region are indicated. The vertical dashed lines indicate the end points of subclones made from the traY-to-traL fragment. Filled and open boxes represent the wild-type and mutant −35 and −10 hexamers of the traY promoter, respectively. pUBF36 contains a 71-bp deletion of the traY promoter. The Ω fragment present in pUBF38Ω1 contains a gene encoding spectinomycin resistance (Spr) and is flanked by transcriptional terminators (T). Insertion specificity is defined as the number of insertions in region I divided by the total number of insertions. The orientation of IS903 insertion is indicated by small arrows (→ and ←). Line 1 shows that 31 insertions were mapped in the same orientation in pUB307 (data from Fig. 4). Transposition frequency was determined from at least three independent mating-out assays.