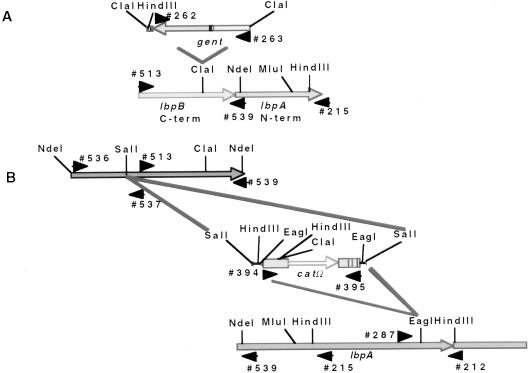
FIG. 3.
(A) Construction of an N. meningitidis N16T12EK lbpB::gent isogenic mutant. The lbpB gene was amplified by PCR using primers 513 and 215 and cloned into pCR2.1. A gentamicin (gent) resistance marker with flanking ClaI sites was ligated into the unique ClaI site. An E. coli colony was isolated with the gent cassette in the orientation opposite to that of the lbpB reading frame (determined by PCR analysis). The DNA was linearized by using unique restriction enzymes sites in the pCR2.1 polylinker sequence, and the DNA was electroporated into N. meningitidis N16T12EK (TbpB− TbpA−). (B) Construction of N. meningitidis N16T12EK lbpB::catΩ and lbpA::catΩ isogenic mutants. The entire N. meningitidis B16B6 lbpB gene was amplified by PCR using primers 536 and 539 and cloned into pCR2.1. A chloramphenicol acetyltransferase omega (catΩ) resistance marker with flanking SalI sites was ligated into the unique SalI site of the B16B6 lbpB gene, and the catΩ with flanking EagI sites was ligated into the unique EagI site of the strain BNCV lbpA gene (37). Orientation of the inserted catΩ gene was determined by PCR analysis. The DNA was linearized by using NdeI sites that flanked the DNA inserts and used for natural transformation into N. meningitidis N16T12EK. The NdeI site at the 5′ end of the B16B6 lbpB gene was incorporated by using site-specific mutagenesis with primer 536 (Table 1). The gent and catΩ cassette insertions are as shown. The catΩ cassette was inserted in the orientation opposite to that of the lbpA gene.