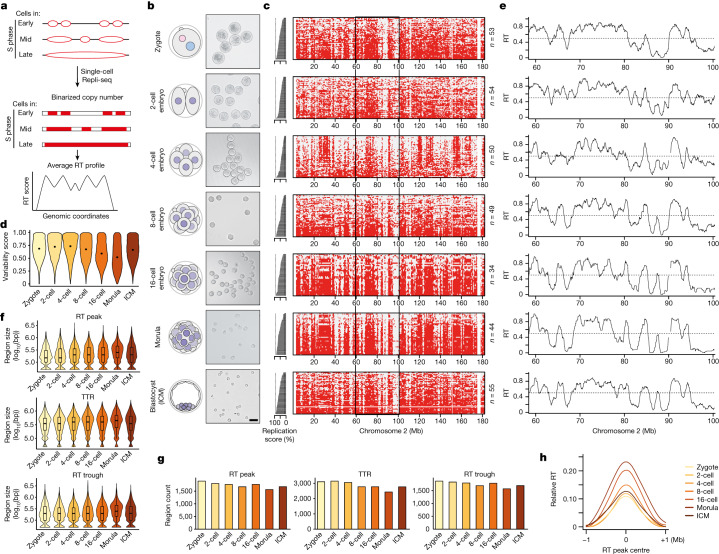
Fig. 1. RT emerges gradually during mouse preimplantation development.
a, Overview of single-cell Repli-seq used to generate RT profiles from single cells in mouse preimplantation embryos based on copy number variation. b, Schematic of sampling of embryos and corresponding images of dissociated blastomeres at each stage. The numbers of independent blastomere collections for each stage with similar results are as follows: zygote (3), 2-cell (4), 4-cell (3), 8-cell (3), 16-cell (3), morula (2), ICM (4). Scale bar, 50 μm. c, Heatmaps of single cells indicating replication status based on binarized copy number during preimplantation embryogenesis (red, replicated; grey, not replicated). Cells are ranked by their percentage of replicated genome (replication score), which indicates progress in S phase and is plotted as a bar plot on the left. d, Variability score during embryonic development; the score is 1 when 50% of cells replicated the genomic bin and 0 when all cells are either replicated (100%) or non-replicated (0%). Each violin plot shows the distribution of scores for all genomic bins. e, RT profiles of preimplantation embryos over a representative region on chromosome 2, denoted by black rectangle in c. Black line indicates RT profiles, calculated as the average of overlapping intervals defined by genome-wide replication score. f,g, Size (f) and number (g) of replication features RT peaks (also known as initiation zones), and RT troughs (also known as termination zones) during preimplantation development. Box plots show median and interquartile range (IQR), and whiskers depict the lowest and highest values within 1.5× IQR. bp, base pair. h, Relative RT values centred at RT peaks during embryonic development compared with their neighbouring regions. Note that curves for the 2- and 4-cell stages overlap considerably and, to some extent, with that of zygotes.