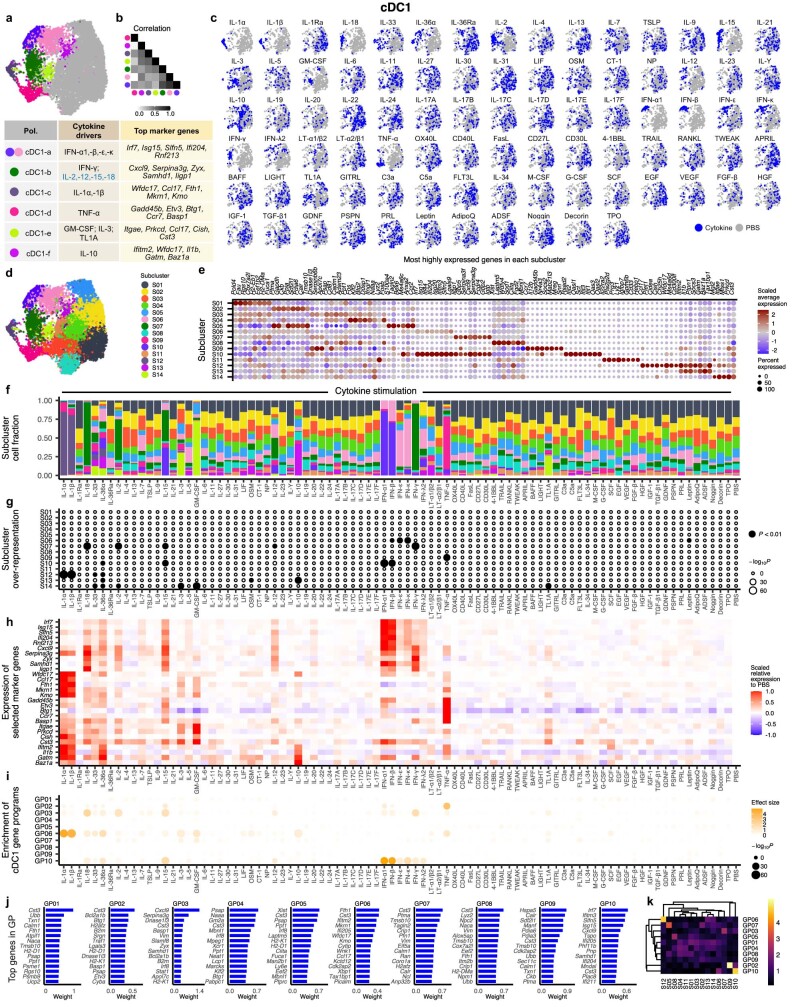
Extended Data Fig. 7. cDC1 responses to cytokines: polarization states, subclusters, marker gene expression, and gene programs.
a, Top, UMAP visualization of cDC1 cells for all cytokines, colored by polarization states; bottom, table with cell type polarization states (left column), single cytokine drivers (middle column), and top marker genes (right column); reproduced from Fig. 3 for ease of reference. b, Pairwise Pearson correlation coefficients between polarization states. c, UMAP visualization of cDC1 cells shown independently for each cytokine treatment, colored by cytokine treatment (blue) or PBS treatment control (gray). d, UMAP visualization of cDC1 cells for all cytokine or PBS treatment; cells colored for cDC1 Louvain subclusters. e, Top overexpressed genes in each Louvain subcluster in d; color, column-scaled average expression; size of circle, percentage of cells in the subcluster expressing each gene. f-i, cDC1 responses to each cytokine stimulation. f, Fraction of cells per subcluster in each cytokine treatment. Colors represent subclusters defined in d. g, Enrichment of each subcluster in each cytokine treatment; size of circle, Bonferroni-adjusted P-value of hypergeometric test relative to PBS; black fills, P < 0.01. h, Row-normalized relative expression of representative marker genes of each polarization state in cytokine-treated vs. PBS-treated cells. i, Enrichment of cDC1 gene programs obtained from NMF analysis of all cDC1 cells in cytokine-treated cells relative to PBS-treated cells; size of circle, FDR-adjusted P-value from two-sided Wilcoxon rank-sum test; shade, effect size representing the mean difference in gene program weight. j, Top weighted genes in each gene program in i. k, Average gene program weight in each subcluster. Rows and columns were hierarchically clustered using the complete-linkage method on Euclidean distances.