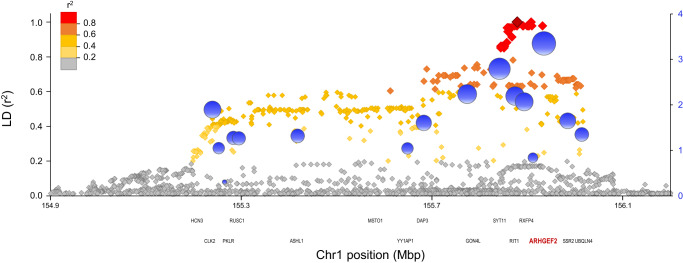
Fig. 8. ARHGEF2 containing IBD risk locus on chromosome 1 (1q22).
The single nucleotide polymorphism (SNP) rs670523 was previously associated to IBD by GWAS44. The graph shows the linkage disequilibrium structure of the proxy SNPs to rs670523 (dark gray) calculated using LDlink64 as r2 values based on the 1000 Genomes phase 3v5 CEU + GBR European reference haplotypes (left y axis). The proxy SNPs are color-coded according to their r2 values and positioned at their chromosomal location (x-axis) over the genes found at this locus. The right y-axis shows the results of the MIS (Myeloid Inflammatory Score) epigenetic scoring method that we previously described6 and designed to prioritize candidate genes involved in inflammatory processes in myeloid cells based on functional genomics information. The MIS for each gene is shown by a blue bubble over its transcriptional start site with a diameter proportional to the MIS. ARHGEF2 is found in strong LD with rs670523 and has the highest MIS within this IBD risk locus.