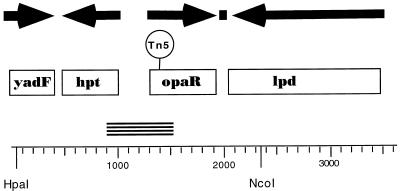
FIG. 2.
Organization of the luxR-like locus of V. parahaemolyticus. The complete double-stranded nucleotide sequence of the region from bp 1 to 3605 was obtained from cosmid pLM1950. Boxes indicate open reading frames (ORFs). Arrows point in the direction of transcription of the deduced coding regions. The solid square shows the location of a potential rho-independent transcriptional terminator. The ORF for yadF (from bp 3 to 411) is incomplete; however, over 136 amino acids its product shows 72% similarity and 63% identity to E. coli YadF by GCG BestFit analysis. The predicted product of hpt (coding region from bp 488 to 1018) is 176 amino acids long and is 99% similar and 95% identical to V. harveyi Hpt. The ORF from bp 1338 to 1952 codes for a 204-amino-acid polypeptide showing 98% similarity and 96% identity to V. harveyi LuxR. The 475-amino-acid deduced product of the ORF from bp 2080 to 3507 exhibits 92% similarity and 87% identity to E. coli Lpd. Tn5 in pLM1952 is contained on a 2.3-kb HpaI-NcoI restriction fragment, and DNA sequencing localized the insertion to bp 1392, which is within the coding region for the LuxR homolog. This gene is designated opaR in V. parahaemolyticus. The PCR product produced with the HPT and LUXUR1 primers is represented by the stacked short lines.