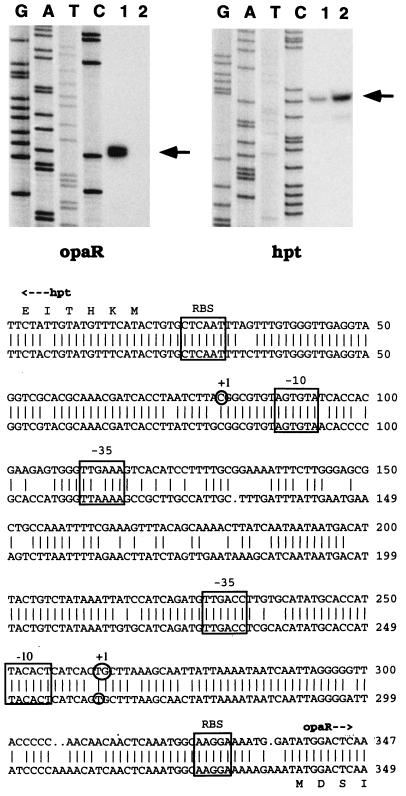
FIG. 7.
Primer extension mapping of opaR and hpt transcripts. RNA was prepared from BB22OP (lane 1) and BB22TR (lane 2). Lanes G, A, T, and C correspond to the sequencing reactions generated with opaR- or hpt-specific primers for pLM1950 template DNA. The positions of the primer extension products are indicated by arrows. Below the autoradiogram is a GCG BestFit comparison of the V. parahaemolyticus (top) and V. harveyi (bottom) intergenic regions spanning the hpt and the opaR (or luxR) genes. Features of the sequence include potential ribosome binding sites identified by homology with the Shine-Dalgarno sequence (RBS), potential ς70 promoters (boxed −10 and −35 regions), and transcription start sites (circled nucleotides labeled +1). For V. harveyi, the luxR start site was determined by Miyamoto et al. (25), and the promoter boxes were proposed by Showalter et al. (29) and Miyamoto et al. (25). For V. parahaemolyticus, a strong doublet was observed for the opaR primer extension product (circle including two bases). The doublet was very clear in autoradiogram exposures shorter than the one shown here. It should be noted that there are multiple potential ATG start codons for opaR, and the translation depicted in this figure represents the particular N terminus having optimal spacing with respect to a potential Shine-Dalgarno sequence and corresponding to the predicted V. harveyi N terminus.