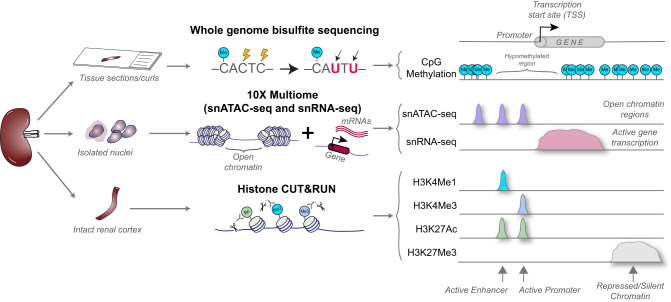
Fig. 1. Study workflow.
In an overlapping set of kidney samples, tissue was interrogated by laser microdissection-guided whole genome bisulfite sequencing (WGBS), by multiome single nucleus Assay for Transposase-Accessible Chromatin for sequencing (snATAC-seq) and single nucleus RNA sequencing (snRNAseq) after cell disaggregation, and by Cleavage Under Targets & Release Using Nuclease (CUT&RUN) of kidney cortex. Datasets were aligned in the human genome 38 to create an integrated epigenomic atlas.