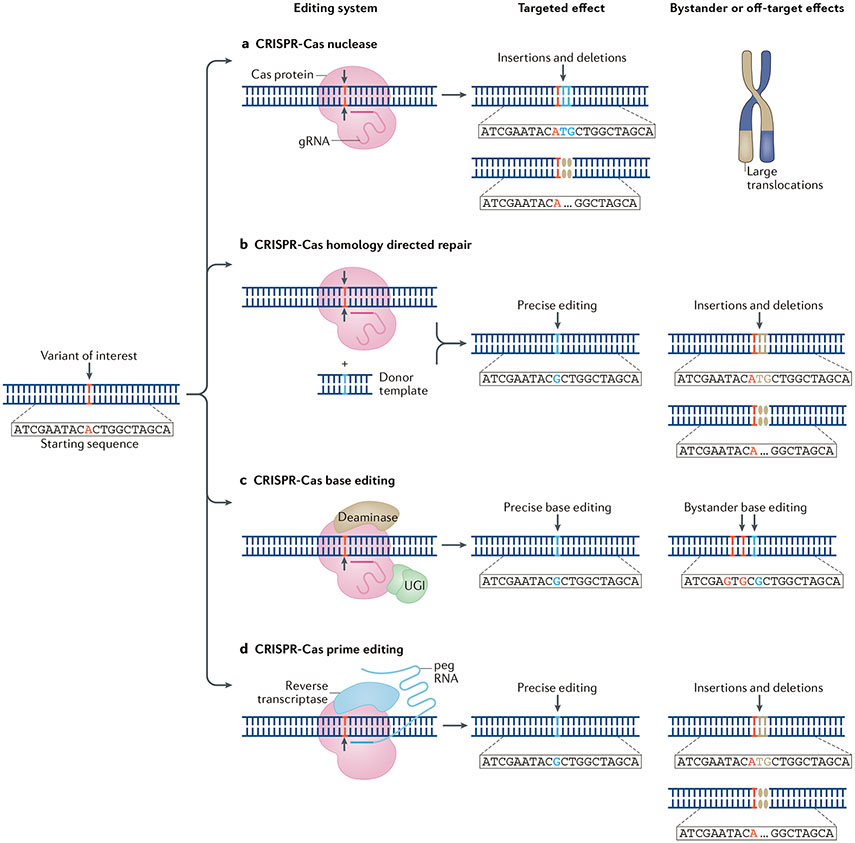
Fig. 3 ∣. Genomic editing to investigate variants of interest in rheumatic disease.
A variant of interest can be investigated using a number of genomic editing techniques. a ∣ The area of interest can be altered with small deletions or insertions using CRISPR–Cas nucleases. b ∣ Targeted homology-directed repair with CRISPR–Cas nucleases and a donor template can be used to precisely edit the variant of interest. c ∣ CRISPR–Cas base editors (comprising a nuclease-deactivated Cas9 protein (dCas9) or Cas nickase fused with an adenine or cytosine deaminase and a uracil DNA glycosylase inhibitor (UGI)) can be used to directly edit a base of interest. d ∣ CRISPR–Cas prime editors (comprising a Cas endonuclease or Cas nickase fused with a reverse transcriptase that is programmed with a prime editing guide RNA (pegRNA)) can be used for precise editing of the target site. For each approach, bystander or unwanted off-target effects can occur (as shown).