Figure 1. Mutation accumulation and antibody neutralization of SARS-CoV-2 S protein.
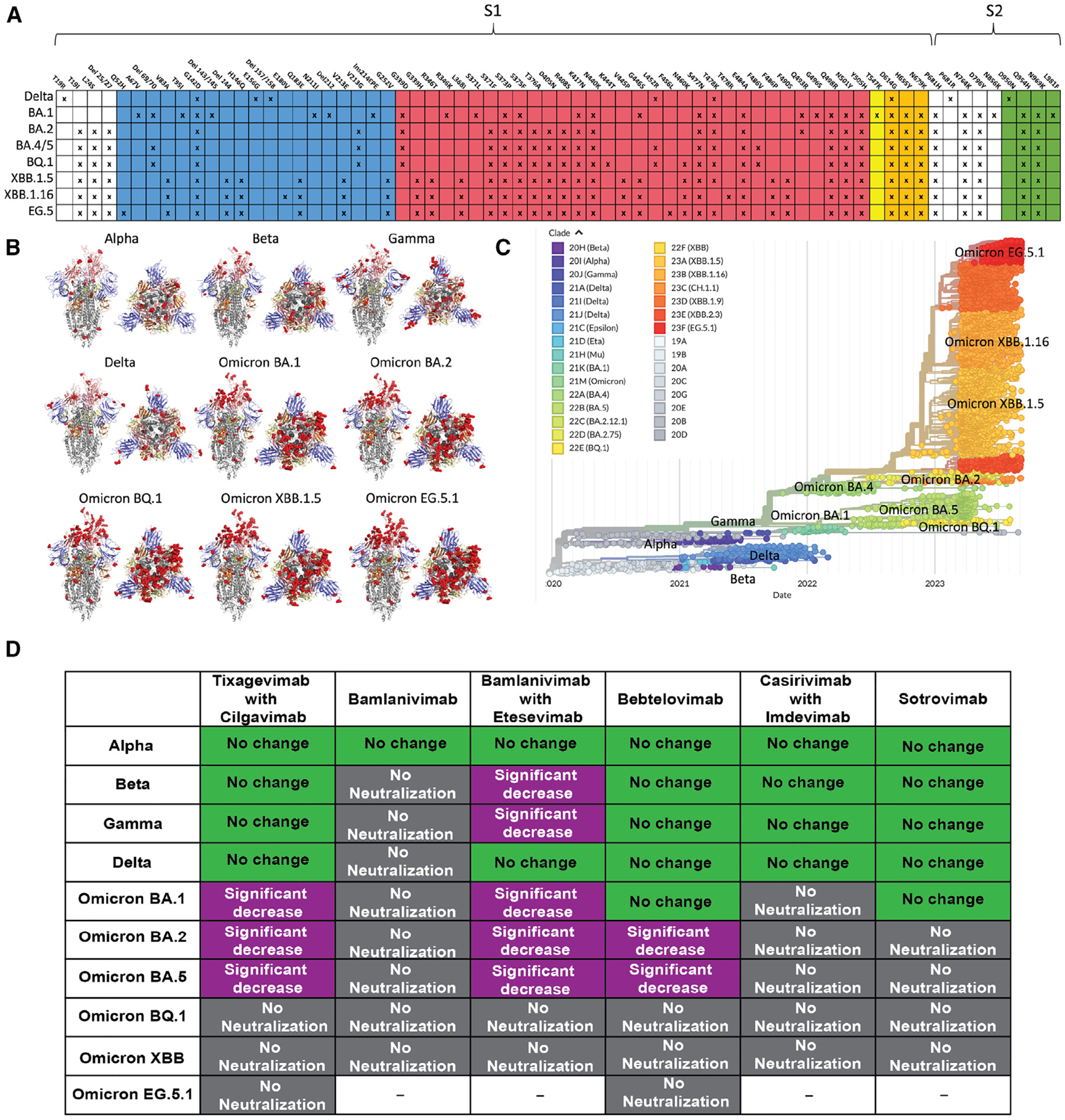
(A) Highest frequency Delta and Omicron SARS-CoV-2 variant mutations relative to the initial strain are indicated with Xs. Blue: NTD mutations; red: RBD mutations; yellow: SD1 mutations; orange: SD2 mutations. These diagrams were created using data from the Lineage Comparison tool from GISAID.
(B) Locations of mutations relative to the initial strain mapped on SARS-CoV-2 variant S protein structures. Mutations are shown by red dots. Mutations shown are >60% prevalent in each variant. S structure coloring follows that of (A) and Figure 2A. All structures are in the 1-up-RBD conformation. Mutations shown on each structure are as follows: Alpha: N501Y, A570D, D614G, T716I, S982A, D1118H; not shown: P681H; Beta: D215G, K417N, E484K, N501Y, D614G, A701V; not shown: D80A; Gamma: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I; and not shown: V1176F. Delta and Omicron mutations are those shown in (A). PDB IDs of S structures are Alpha, PDB: 7EDF; Beta, PDB: 7LYQ; Gamma, PDB: 8DLO; Delta, PDB: 7V7P; and Omicron BA.1, PDB: 7TL9. BA.2, BQ.1, XBB.1.5, and EG.5.1 mutations are all modeled on PDB: 7TL9.
(C) Phylogenetic tree showing the evolution of Omicron over time. Generated from GISAID.
(D) FDA-approved monoclonal antibodies for treatment of SARS-CoV-2 and their efficacy against VOCs and Omicron variants in comparison to the initial SARS-CoV-2 strain. Green: no change; purple: significant decrease; gray: no neutralization. Data are sourced from Qu et al.,20 Tao et al.,23 Imai et al.,30 and Wang et al.,31 as well as https://www.fda.gov/media/145611/download, https://www.fda.gov/media/145802/download, https://www.fda.gov/media/149534/download, https://www.fda.gov/media/154701/download, https://www.fda.gov/media/156152/download, https://covdb.stanford.edu/susceptibility-data/table-mab-susc/, and https://www.bv-brc.org/view/VariantLineage/.
