Figure 3. The compact nature of the Omicron S protein compared to previous SARS-CoV-2 variants.
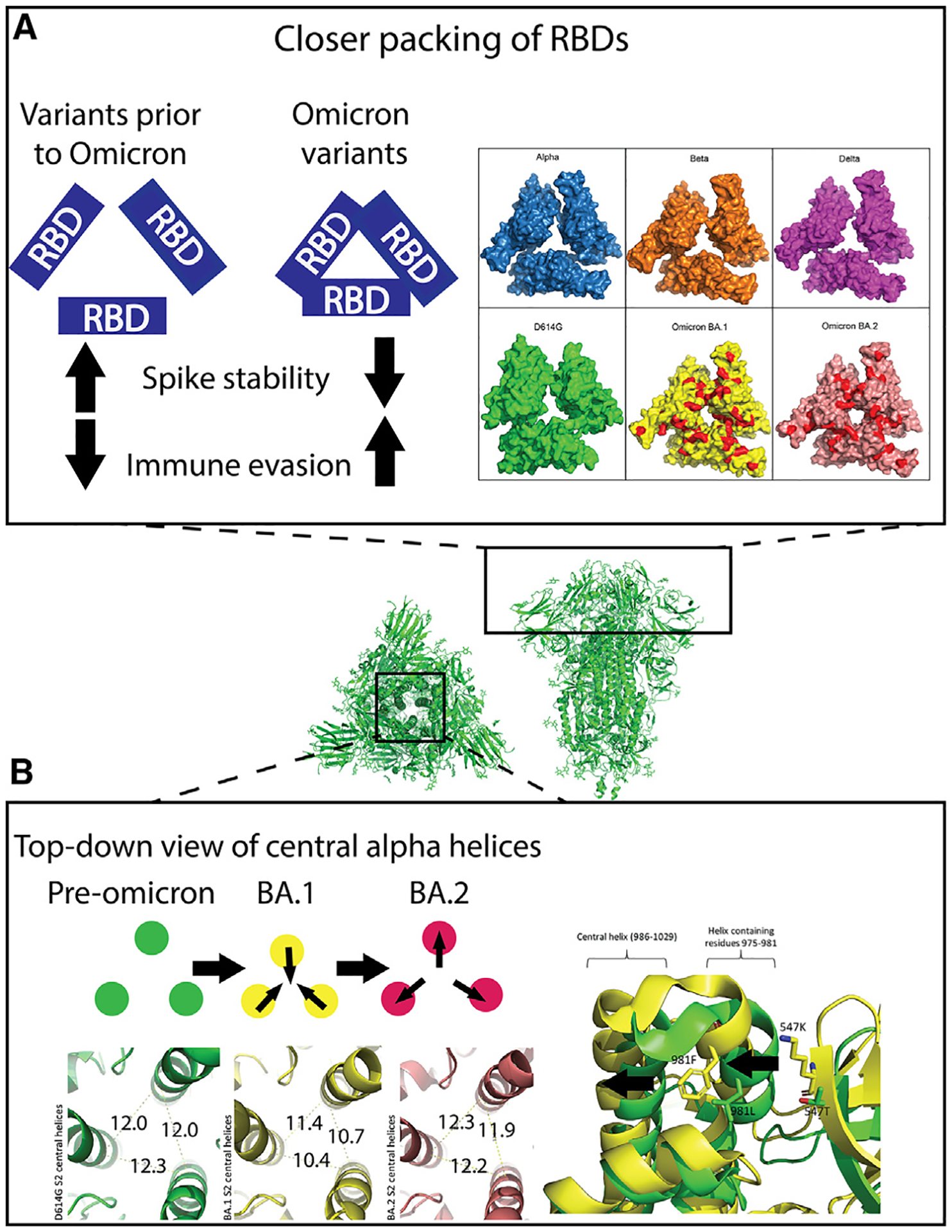
Center: top and side views of the 3-RBD-down D614G S ectodomain (PDB: 7KE8).
(A) Left: trend in Omicron variants to optimize inter-RBD contacts leading to stabilization of the RBD-down state, resulting in higher immune evasion. Right: map of RBD mutations on solved closed RBD structures in surface view. PDB IDs: Alpha, PDB: 7R13; Beta, PDB: 7LYL; Delta B.1.617.2, PDB: 7TOU; D614G, PDB: 7KDK; BA.1, PDB: 7TL1; BA.2, PDB: 7UB6. Red patches on BA.1 and BA.2: sites of mutation in Omicron variants relative to Delta variant.
(B) Top left: schematic showing the inward and outward movements of the S S2 central helices in pre-Omicron, BA.1 and BA.2. Bottom left: green, D614G S2 central helix (PDB: 7KE8) distances measured from C alpha-atom of residue R995 on each monomer; yellow: Omicron BA.1 S2 central helix (PDB: 7TF8) distances measured the same as D614G; red: Omicron BA.2 S2 central helix (PDB: 7UB0) distances measured the same as above. Right: aligned structures of D614G (green) PDB: 7KE8 and BA.1 (yellow) PDB: 7TF8. Residues 547 and 981 are shown as sticks. The arrows indicate the movements of the two helices due to these two mutations.
