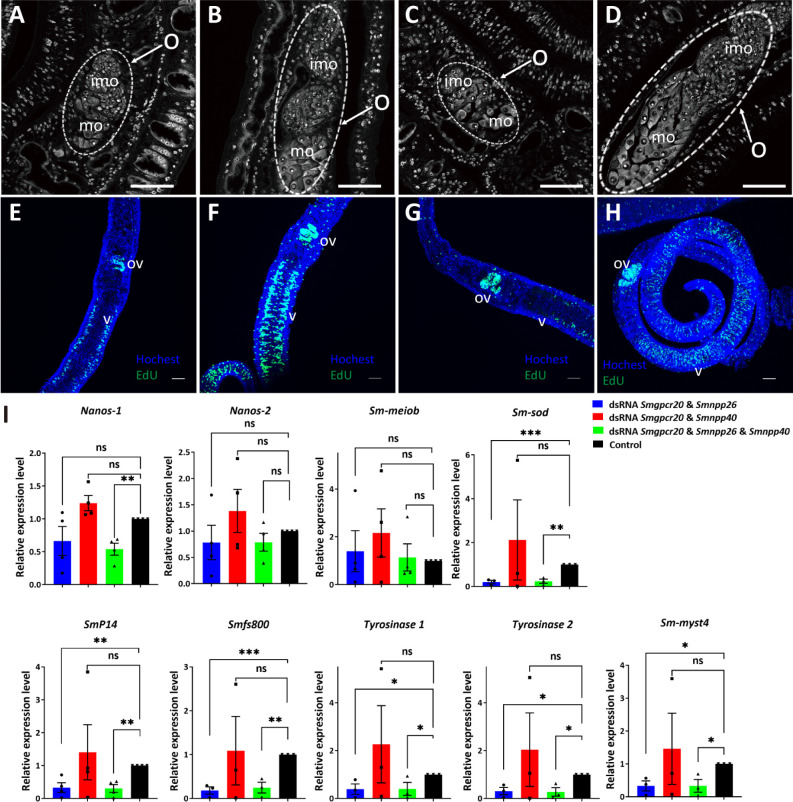
Fig 9.
RNAi against Smgpcr20, Smnpp26, and Smnpp40 affected gonad development, oocyte differentiation, and gene expression in first-time paired S. mansoni females. CLSM analysis showing representative pictures of S. mansoni females post pairing for 21 days (see Fig. 9A) in the presence of Smgpcr20/Smnpp26 dsRNA (A), Smgpcr20/Smnpp40 dsRNA (B), or a combination of Smgpcr20/Smnpp26/Smnpp40 dsRNAs (C). First-time paired females maintained under the same conditions but without dsRNA served as control (D) (n = 5). O, ovary; imo, immature oocytes; mo, mature oocytes; scale bars: 50 µm. (E–H) Exemplary results of EdU staining of paired females (five each per experiment) following dsRNA treatment; Hoechst 33342 was used as counterstaining (blue), ov, ovary; v, vitellarium. The order of E–H is as in A–D. Compared to the control (H), a reduction of the number of EdU-stained cells was mainly observed in the Smgpcr20/Smnpp26 (A) and the triple dsRNA treatment groups (C). (I) qRT-PCR experiments were performed to determine the transcript levels of selected genes. Fold changes of gene expression levels were calculated using the 2−∆∆Ct method. The transcript levels of genes encoding nanos-1, nanos-2, Smp14, fs800, Smtyr1, Smtyr2, Smmyst4, and Smsod were significantly lower than in the control group; nanos-2 and Smmeiob transcript levels showed no significant differences among the RNAi and the control groups (n >3). Data are representatives of the mean ± SEM of three separate experiments. Significant differences determined by t-test were indicated as: ****P < 0.0001, **P < 0.01, *P < 0.05.