In Brief
The authors performed a functional genomics analysis of nonsyndromic craniosynostosis (nsCS) to determine when, where, and in what cell types nsCS-associated genes converge during development. They found that nsCS genetic risk converges in cells and transcriptional networks that concurrently impact skull, meninges, and brain development. These findings suggest a primary impact on neurodevelopment in nsCS, providing an explanation for why some children with nsCS continue to have neurodevelopmental disability despite correction of cranial defects. The authors hypothesize that genetic factors may be more important for determining overall neurodevelopmental outcome than either the timing or type of surgical correction used (endoscopic vs open).
Keywords: craniosynostosis, autism, neurodevelopmental disorders, integrative genomics, cerebral cortex, cortical development, craniofacial
ABBREVIATIONS : CGE = caudal ganglionic eminence, CS = craniosynostosis, DD = developmental disorder, DNM = de novo mutation, FDR = false discovery rate, GO = Gene Ontology, IN-CTX-CGE = CGE-derived inhibitory neuron, IN-CTX-MGE = MGE-derived inhibitory neuron, IPC = intermediate progenitor cell, LOF = loss of function, MGE = medial ganglionic eminence, ncWNT = noncanonical WNT, nsCS = nonsyndromic CS, OG = osteogenic cluster, PCW = postconception week, PO = proliferative osteogenic cluster, rWGCNA = robust consensus WGCNA, scNRA-seq = single-cell RNA sequencing, WES = whole-exome sequencing, WGCNA = weighted gene coexpression network analysis.
Abstract
OBJECTIVE
Nonsyndromic craniosynostosis (nsCS), characterized by premature cranial suture fusion, is considered a primary skull disorder in which impact on neurodevelopment, if present, results from the mechanical hindrance of brain growth. Despite surgical repair of the cranial defect, neurocognitive deficits persist in nearly half of affected children. Therefore, the authors performed a functional genomics analysis of nsCS to determine when, where, and in what cell types nsCS-associated genes converge during development.
METHODS
The authors integrated whole-exome sequencing data from 291 nsCS proband-parent trios with 29,803 single-cell transcriptomes of the prenatal and postnatal neurocranial complex to inform when, where, and in what cell types nsCS-mutated genes might exert their pathophysiological effects.
RESULTS
The authors found that nsCS-mutated genes converged in cranial osteoprogenitors and pial fibroblasts and their transcriptional networks that regulate both skull ossification and cerebral neurogenesis. Nonsyndromic CS–mutated genes also converged in inhibitory neurons and gene coexpression modules that overlapped with autism and other developmental disorders. Ligand-receptor cell-cell communication analysis uncovered crosstalk between suture osteoblasts and neurons via the nsCS-associated BMP, FGF, and noncanonical WNT signaling pathways.
CONCLUSIONS
These data implicate a concurrent impact of nsCS-associated de novo mutations on cranial morphogenesis and cortical development via cell- and non–cell-autonomous mechanisms in a developmental nexus of fetal osteoblasts, pial fibroblasts, and neurons. These results suggest that neurodevelopmental outcomes in nsCS patients may be driven more by mutational status than surgical technique.
The neurocranial complex, comprising the cranium, meninges, and brain, develops "hand in glove,"1 with each component influencing the development of the others via poorly understood mechanical and molecular signals.1,2 One paradigm that illustrates the importance of bone-brain interactions during development is craniosynostosis (CS), the most prevalent skull deformity in children, affecting approximately 1 in 2000 live births.3,4 CS is characterized by the premature fusion of one or more sutures of the developing calvaria that can hinder the expansion of the growing brain. Most CS cases present as an isolated defect, termed nonsyndromic CS (nsCS). The treatment for CS is surgical repair to restore the normal appearance of the skull and prevent damaging elevations in intracranial pressure.5 Despite optimal surgical correction, long-term neurodevelopmental sequelae can persist in up to half of patients.6,7
Recent investigations have expanded our knowledge of the genetic causes of nsCS, with results implicating distinct processes involved in suture formation such as abnormal osteoprogenitor proliferation and function.8–12 Whole-exome sequencing (WES) of sporadic nsCS trios has discovered rare, loss-of-function (LOF), and damaging missense mutations with large effect size.9,10 Despite these advancements in gene discovery, the mechanisms linking gene mutations to disease pathology, especially with regard to neurodevelopmental outcomes, remain poorly understood.
In this study, we asked when, in which brain regions, and in what cell types genes with nsCS-associated de novo mutations (DNMs) converge during neurocranial development. Based on an emerging concept suggesting that heterogeneous genetic loci converge on specific cellular processes in neurodevelopmental disorders,13–15 we hypothesized that DNMs in biologically pleiotropic genes disrupt a nexus of intersecting processes at the prenatal cranium, meninges, and brain to drive both premature cranial suture closure and intrinsic brain pathology in nsCS. To test this hypothesis, we mapped mutation-intolerant genes harboring rare, LOF, and damaging missense nsCS-associated DNMs defined by unbiased WES data from 291 nsCS proband-parent trios10 onto genome-wide bulk and single-cell RNA sequencing (scRNA-seq) data sets of the developing mouse and human cranial sutures,16 meninges,17 and brain.18,19
We found that genes harboring nsCS-associated DNMs converged not only on fetal cranial osteoprogenitors and their processes involved in connective tissue biology and suture maturation but also on discrete populations of inhibitory neurons and transcriptional networks previously implicated in autism and other neuropsychiatric disorders. Using cell-cell communication analysis, we identified bidirectional crosstalk between osteoblasts and neurons of the fetal brain via multiple ncCS-related signaling pathways. These findings provide a compelling theory of how genetic lesions concurrently impacting cranial morphogenesis and neurodevelopment in nsCS elicit persistent neurocognitive impairment despite optimal surgical repair of the cranial vault.
Methods
Disease Cohort and Determination of Disease Risk Gene Lists
The study is based on publicly available genetic and transcriptomic data sets.16–19 As described previously,10 participants in the original study were from the Yale Pediatric Craniofacial Clinic, the Pediatric Neurosurgery Clinic at the Medical University of Silesia, Poland, or through invitations from Cranio Kids–Craniosynostosis Support and Craniosynostosis–Positional Plagiocephaly Support Facebook pages. Those diagnosed with sagittal or metopic CS and no known syndromic forms of disease by a craniofacial plastic surgeon or pediatric neurosurgeon were included. Saliva or buccal swab samples were obtained from families. Approval for the study protocol was granted by the Yale Human Investigation Committee Institutional Review Board.
Risk genes from our nsCS cohort were defined as genes that harbor at least one de novo protein-altering mutation identified by an exome sequencing study of nsCS with 291 parent-offspring trios.10 Based on these criteria, the nsCS risk gene list consisted of 86 genes (Table S1).10 Risk genes for autism (Table S2),20,21 developmental disorder (DD) (Table S3),22 schizophrenia (Table S4),23 and height (Table S5)24 were extracted from the literature as previously described.
Developing Human Brain, Mouse Meninges, and Suture scRNA-seq Data Set Analyses
As described previously,16,17,19 the preprocessing and clustering analyses for developing brain, meninges, and suture data sets were completed using Seurat.25 Briefly, cells with a low number of unique molecular identifiers, genes, or high mitochondrial content were filtered. Cell counts were normalized, and cell-cycle regression was performed. Dimensionality reduction and clustering were performed using FindNeighbors and FindClusters, and marker genes were identified with FindAllMarkers. Subclustering was performed as described above. Main and subcluster annotation was performed based on canonical markers as described previously.16,17,19
Clusters for developing human brain were assigned to cells based on previous analysis via a hybrid method using Louvain clustering and weighted gene coexpression network analysis (WGCNA).19 FindAllMarkers was used to identify differentially expressed markers across time points, areas, and laminar zones by running. Heatmap expression values were calculated using the AverageExpression function, and visualization of the heatmaps was created using the pheatmap package.
Gene Coexpression Network Creation From scRNA-seq Data Sets
The gene coexpression modules for the scRNA-seq data sets were created through the differential expression analysis workflow of Monocle3.26 Monocle3’s graph_test(neighbor_graph="knn") function was applied to identify genes with varying expression between cell clusters.26 Genes with a false discovery rate (FDR)–adjusted p value < 0.05 were used to create gene coexpression networks. The R package hypeR27 was used to complete hypergeometric enrichment and identify modules significantly enriched (FDR-adjusted p value < 0.05) with nsCS-associated genes.
CellChat Analysis
The official CellChat28 pipeline was used to infer intercellular communication networks. Briefly, the functions identifyOverExpressedGenes, identifyOverExpressedInteractions, and projectData were applied to the normalized counts to preprocess the data. The main analyses were then completed via the computeCommunProb, computeCommunProbPathway, and aggregateNet functions using fixed randomization seeds.
Mapping nsCS Genes to Gene Coexpression Networks of the Developing Human Brain
Standard workflow of robust consensus WGCNA (rWGCNA)29 was applied to a processed bulk mRNA-seq data set18 across 16 human brain regions during development between postconception week (PCW) 9 and postnatal year 3. Briefly, samples exceeding 3 standard deviations above the mean sample network connectivity were eliminated. Network analysis was conducted with rWGCNA using gene biweight midcorrelations. To attain scale-free topology (r2 > 0.9), a soft threshold power of 10 was selected. A topological overlap dendrogram was used to generate modules with parameters of minimum module size of 40, deep split of 4, and merge threshold of 0.1.
Indicator-based enrichment was assessed via logistic regression, including all genes present in coexpression modules as background: is.disease ~ is.module + gene covariates (GC [guanine-cystine] content, gene length, and mean expression in bulk RNA-seq atlas).30 Of the 88 WGCNA modules, the gray module consists of genes unassigned to a coexpression network by WGCNA convention29 and thus was excluded from analysis; p values were adjusted by FDR correction.
Cell-Type Enrichment Analysis
Cell-type–enriched genes (cell-type markers) were obtained for each of the developing brain, meninges, and suture scRNA-seq atlases. In a background set of all genes expressed in ≥ 3 cells of each scRNA-seq atlas, logistic regression was applied for indicator-based enrichment analysis: is.cell.type ~ is.disease + gene covariates (GC content, gene length).30 All p values were adjusted by FDR correction.
Gene Ontology Enrichment Analysis
Gene Ontology (GO; biological processes) was performed using the Enrichr R package.31 The top 10 terms with the lowest p values were reported; p < 0.05 was considered significant.
Results
Genes With nsCS-Associated DNMs Are Enriched in Cell Types and Pathways Implicating Mesenchyme-Neuronal Crosstalk
To perform an integrative genomics analysis of nsCS genetic risk with the goal of clarifying disease mechanisms, we ascertained a heterogeneous collection of 86 genes from a WES study of 291 nsCS patient-proband trios (Fig. 1, Table S1).10 We limited input genes to those discovered via hypothesis-naive exome-wide sequencing and applied consistent statistical thresholds for inclusion, allowing us to reduce the potential biases that could arise from attempting to elucidate mechanisms using genes selected, at least in part, based on their biological plausibility.14 Included nsCS-mutated genes were those harboring at least one de novo LOF or D-mis mutation as predicted by MetaSVM.10 Given the results of other integrative genomics studies,13,14,18,32 we reasoned that mapping the convergence of nsCS-mutated genes onto transcriptomic atlases of major structures within the neurocranial complex (bone, meninges, and brain) might enable a relatively unbiased designation of the biological processes affected by nsCS-associated de novo gene mutations with probable pathogenicity (Fig. 1).
FIG. 1.
Schematic illustration of the multiomics integrative genomics analysis pipeline. PY = postnatal year.
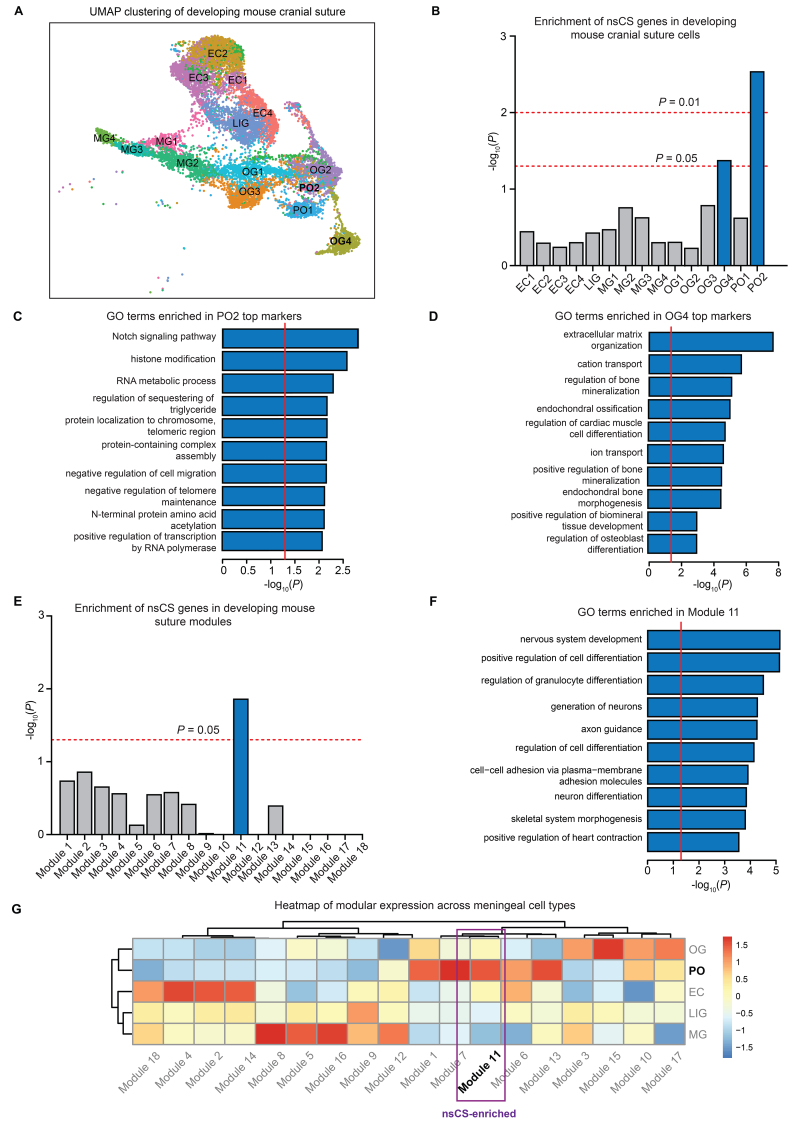
Previous work implicated the abnormal growth and proliferation of suture osteoprogenitors as a developmental mechanism underlying premature suture fusion in CS.8,12 Consistent with their probable role in disease pathophysiology, GO pathway analysis of nsCS-mutated genes revealed enrichment in multiple terms related to mesenchymal development, including regulation of focal adhesion assembly, regulation of cell-matrix adhesion, Wnt signaling, and fibroblast growth factor receptor signaling (Fig. S1). We next mapped nsCS-mutated genes to cell types in an scRNA-seq transcriptomic atlas of the developing mouse coronal suture at E15–17 (i.e., embryonal age of 15–17 days) composed of 16,961 total cells (Fig. 2A).16 We constructed modules of coexpressed genes that we then mapped to specific cell-type clusters. These were then assessed for the enrichment of nsCS-mutated genes and other disease genes (see Methods). As expected, we found enrichment of nsCS-mutated genes in two cell clusters: osteogenic cluster 4 (OG4; p = 4.66 × 10−2) and proliferative osteogenic cluster 2 (PO2; p = 6.3 × 10−3) (Fig. 2B). The top cell-type markers for OG4 and PO2 were enriched for GO terms related to epigenetic regulation such as histone modification (PO2) and terms related to mesenchyme development including negative regulation of cell migration (PO2), extracellular matrix organization (OG4), and regulation of bone mineralization (OG4) (Fig. 2C and D, Fig. S2).
FIG. 2.
Single-cell RNA-seq developing suture data set analysis reveals nsCS enrichment in osteoblasts and neurodevelopmental processes. A: Uniform manifold approximation and projection (UMAP) clustering of developing mouse coronal suture cells. B: Enrichment of nsCS risk genes in developing mouse cranial suture cell types. Analyzed transcriptomic data set from Farmer et al.16 Logistic regression for indicator-based enrichment was used to calculate FDR-adjusted p values. C: GO biological process pathway analysis for top genes in nsCS-enriched proliferative osteoblasts. Red line indicates threshold p value of 0.05. D: GO biological process pathway analysis for top genes in nsCS-enriched mature osteoblasts. Red line indicates threshold p value of 0.05. E: Enrichment of nsCS risk genes in developing mouse suture gene coexpression modules. Analyzed transcriptomic data set from Farmer et al.16 The hypergeometric enrichment test was used to calculate FDR-adjusted p values. F: GO biological process pathway analysis for top genes in nsCS-enriched module 11. Red line indicates threshold p value of 0.05. G: Heatmap of gene coexpression modules across individual cell types in developing mouse suture. EC = ectocranial mesenchymal cluster; LIG = ligament-like mesenchymal cluster; MG = meningeal cluster.
After constructing gene coexpression modules from the scRNA-seq data, we found that nsCS-mutated genes were enriched only in module 11 (p = 1.3 × 10−2) (Fig. 2E). The top terms found in GO pathway analysis showed that the gene coexpression network module 11 regulates not only mesenchymal differentiation through skeletal system morphogenesis and cell-cell adhesion via plasma-membrane adhesion molecules, but also cerebral development via pathways related to neuron differentiation, guidance of axons, and generation of neurons (Fig. 2F, Fig. S3). Despite being most significantly involved in neurodevelopmental processes, nsCS-enriched module 11 showed the highest enrichment in the proliferative osteogenic cluster (PO) (Fig. 2G), implying a link between osteoblasts and brain development.
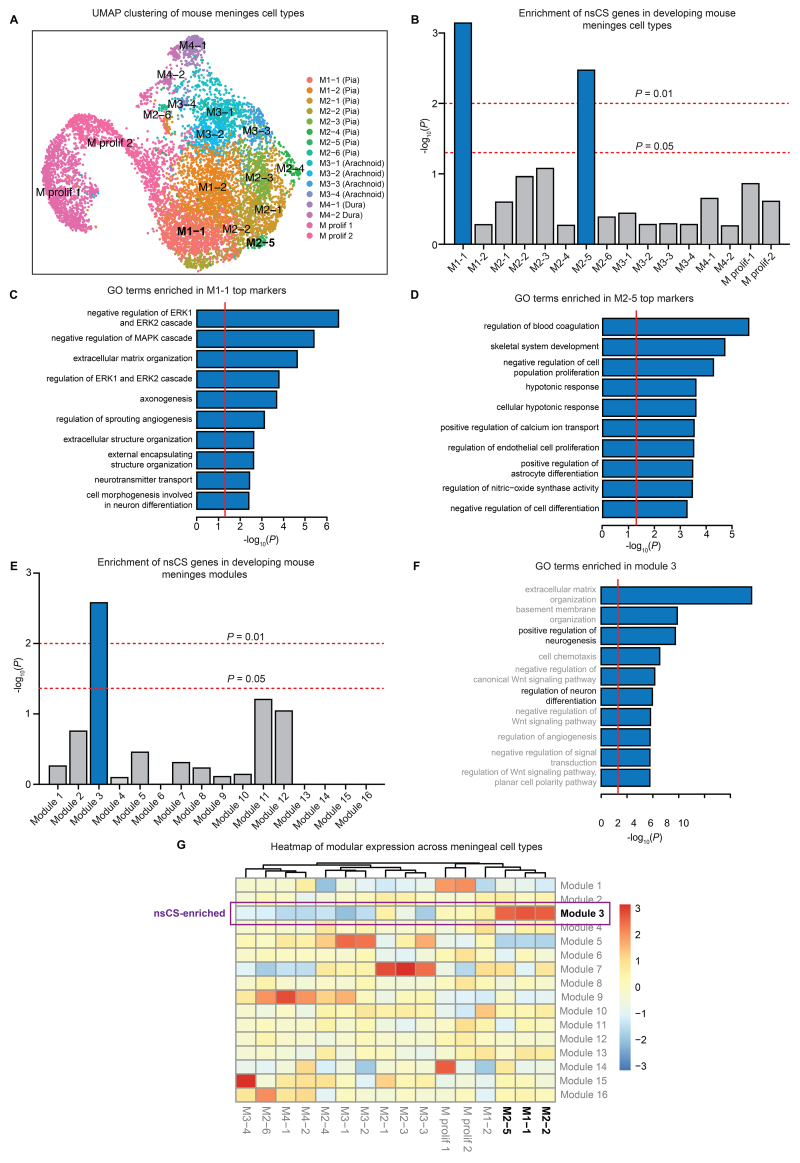
Next, we examined the expression of nsCS-mutated genes in an scRNA-seq transcriptomic data set of E14 developing mouse meninges,17 membranous layers that interact with the overlying skull and underlying brain.17 This data set included FACS (fluorescence-activated cell sorting)–sorted 8851 COLA1+ fibroblasts from all three layers of the embryonic forebrain meninges (Fig. 3A, Fig. S4). The nsCS-mutated genes were enriched in two pial fibroblast subclusters labeled as M1-1 (i.e., subcluster 1 of meninges 1) (p = 7.2 × 10−4) and M2-5 (p = 3.2 × 10−3) (Fig. 3B). Pial fibroblasts form a basement membrane by secreting critical extracellular matrix proteins, and ablation of this basement membrane leads to abnormal brain development by disrupting neuronal migration and proliferation in the neocortex.33 Consistent with this, the top cell markers for nsCS-enriched M1-1 and M2-5 subclusters were enriched with terms related to neurogenesis-related processes such as axonogenesis (M1-1) (Fig. 3C, Fig. S5), cell morphogenesis involved in neuron differentiation (M1-1) (Fig. 3C, Fig. S5), and positive regulation of astrocyte differentiation (M2-5) (Fig. 3D, Fig. S5), and synaptic development processes such as neurotransmitter transport (M1-1) (Fig. 3C) and positive regulation of calcium ion channel transport (M2-5) (Fig. 3D, Fig. S5), in addition to extracellular matrix processes such as extracellular structure organization (M1-1) (Fig. 3C, Fig. S5).
FIG. 3.
Single-cell RNA-seq developing meninges data set analysis reveals nsCS enrichment in pial fibroblasts and synaptic/neuronal differentiation processes. A: UMAP clustering of developing mouse meninges cells. B: Enrichment of nsCS risk genes in developing mouse meninges cell types. Analyzed transcriptomic data set from DeSisto et al.17 Logistic regression for indicator-based enrichment was used to calculate FDR-adjusted p values. C: GO biological process pathway analysis for top genes in nsCS-enriched M1-1 (pial fibroblasts). Red line indicates threshold p value of 0.05. D: GO biological process pathway analysis for top genes in nsCS-enriched M2-5 (pial fibroblasts). Red line indicates threshold p value of 0.05. E: Enrichment of nsCS risk genes in developing mouse meninges gene coexpression modules. Analyzed transcriptomic data set from DeSisto et al.17 The hypergeometric enrichment test was used to calculate FDR-adjusted p values. F: GO biological process pathway analysis for top genes in nsCS-enriched module 3. Red line indicates threshold p value of 0.05. G: Heatmap of gene coexpression modules across individual cell types in developing mouse meninges. M prolif 1 = proliferating meninges 1; M prolif 2 = proliferating meninges 2.
Next, we organized the developing mouse meninges transcriptome into 16 modules of coexpressed genes (see Methods) and identified enrichment of nsCS-mutated genes in one specific module: module 3 (p = 1.5 × 10−2) (Fig. 3E). In addition to enrichment of GO terms related to connective tissue such as extracellular matrix organization and basement membrane organization, module 3 is enriched for terms implicating brain development including positive regulation of neurogenesis and regulation of neuron differentiation (Fig. 3F, Fig. S4). Consistent with cell-type enrichment analysis, this gene coexpression network showed highest expression of nsCS-enriched pial fibroblast subclusters M1-1 and M2-5 (Fig. 3G).
CellChat Analysis Uncovers Significant Ligand-Receptor Interactions Between Osteoblasts and Immature Neurons via the BMP, FGF, and Noncanonical WNT Signaling Pathways
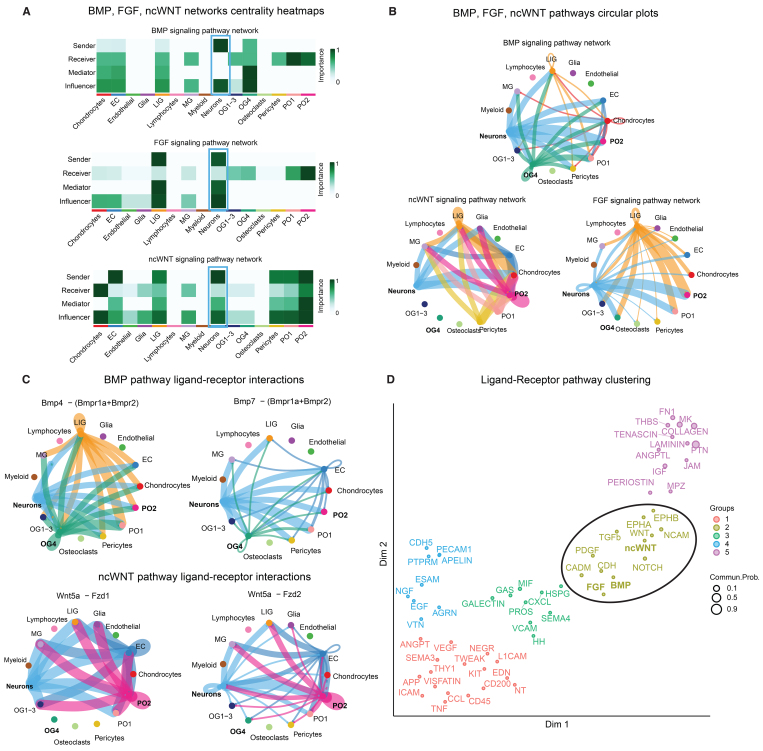
The enrichment of neurodevelopmental and synaptic processes in nsCS-enriched gene modules suggests that genetically impaired mesenchymal tissue may influence cortical development in nsCS. To test this hypothesis, we characterized the crosstalk between connective tissue and neurons in an scRNA-seq transcriptome from the mouse coronal suture16 using CellChat, a bioinformatics tool that uses the expression patterns of known ligand-receptor pairs in cell clusters to infer intercellular communication networks.28 We found that the BMP, FGF, and noncanonical WNT (ncWNT) signaling pathways, which contain genes with multiple nsCS-associated DNMs (SMAD6, SMURF1, DVL3, PSMC2, PSMC5, SPRY1, and SPRY4) (Table S1), were the top three pathways associated with neurons (Fig. 4A). Interestingly, the most significant interactions for these pathways occurred between neurons and nsCS-enriched osteogenic cell clusters (PO2 and OG4) (Fig. 4B).
FIG. 4.
Single-cell RNA-seq CellChat analysis reveals crosstalk between neurons, meninges, and nsCS-enriched osteoblasts at the developmental coronal suture. A: Heatmap showing the relative importance of each cell type from developing mouse coronal suture. Analyzed transcriptomic data set from Farmer et al.16 for BMP, FGF, and ncWNT signaling networks. B: Circle plots showing BMP, FGF, and ncWNT pathway ligand-receptor interactions among cell groups for developing mouse coronal suture. Edge width represents communication strength. C: Circle plots showing individual BMP and ncWNT pathway ligand-receptor pair communication strength among cell groups for developing mouse coronal suture. Edge width represents communication strength. D: Clustering of all significant signaling pathways from developing mouse coronal suture into a shared 2D manifold according to their functional similarity.
Through cell-cell communication analysis of the BMP pathway, we found that the ligand-receptor pairs between Bmp4 (Bmpr1a+Bmpr2) and Bmp7 (Bmpr1a+Bmpr2) contributed most significantly to the BMP osteoblast/neuron communication (Fig. 4C). Interestingly, both Bmp4 and Bmp7 are crucial for both neurulation and the neuronal migration necessary for cerebral cortex development.33 Similarly, further analysis of the ncWNT pathway showed Wnt5a–Fzd1 and Wnt5a–Fzd2 to be the most significant ligand-receptor pairs for ncWNT osteoblast/neuron communication (Fig. 4C). Wnt5a is known to play a crucial role for cortical axon outgrowth in neuroprogenitors as well as osteogenic differentiation in mesenchymal stem cells.34
Next, we conducted unsupervised clustering to identify pathways with high functional similarity based on their interaction patterns among different cell populations in our data set (see Methods). As expected, the FGF, BMP, and ncWNT pathways all clustered together along with the TGF-β and other extracellular matrix organization pathways (Fig. 4D), suggesting interplay among multiple pathways to regulate mesenchymal-neuronal interactions. These findings confirm previously described associations between CS pathogenesis and abnormal growth of mesenchyme8 and suggest that nsCS DNMs disrupt intercellular communication between mesenchymal osteogenic cells and ectodermal neural cells.
Nonsyndromic CS–Mutated Genes Converge in Inhibitory Neurons and Coexpression Networks of the Fetal Human Brain That Overlap With Autism and Other DDs
Given that nsCS gene mutations may influence neurodevelopment indirectly by perturbing the growth of osteogenic cells that communicate with neurons, we wondered whether nsCS genes may have cell-autonomous functions within the brain parenchyma. We therefore analyzed the expression of nsCS-mutated genes in both large-scale bulk and single-cell transcriptomic atlases of normally developing human brains covering the entire lifespan of human development.18,19 Given the increased risk of neuropsychiatric disorders in syndromic CS,35 we also compared nsCS genes with DD genes (Table S2), autism genes (Table S3), and schizophrenia genes (Table S4) (see Methods).
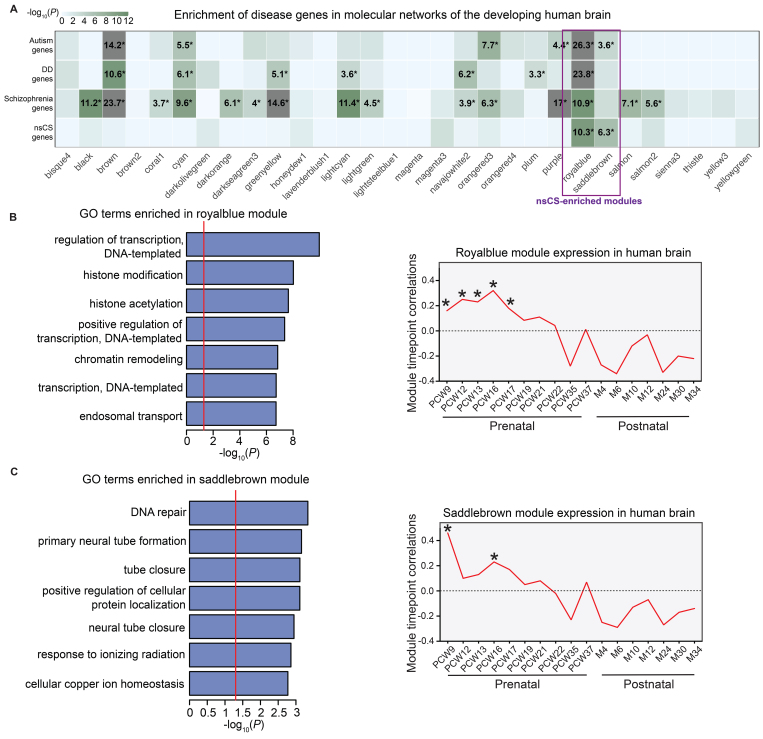
We mapped the various disease risk genes onto molecular networks constructed by WGCNA of human brain RNA sequencing data from 432 histologically verified primary tissue samples from PCW 9 to postnatal year 3 (Fig. S6).18 Each molecular network represents a set of coexpressed genes that share distinct biological functions and/or coregulatory relationships during human brain development,13 allowing us to identify critical pathways during human brain development in which a heterogeneous collection of nsCS risk genes may converge. Among the 87 WGCNA gene coexpression modules generated, nsCS risk genes were enriched in two distinct modules: the "royalblue" module (p = 4.5 × 10−11) and the "saddlebrown" module (p = 4.8 × 10−7) (Fig. 5A). Interestingly, the royalblue module is enriched for schizophrenia (p = 1.2 × 10−11), autism (p = 5.5 × 10−27), and DD (p = 1.5 × 10−24) genes. The saddlebrown module is also enriched for autism risk genes (p = 2.6 × 10−4) (Fig. 5A). As a negative control, genes related to human height (Table S5) are not enriched in any of the 87 WGCNA gene coexpression modules.
FIG. 5.
Convergence of nsCS genes in gene coexpression networks during human brain development in a bulk RNA data set. A: Module-level enrichment for disease risk genes. Modules were constructed via WGCNA of BrainSpan data (see Methods). Logistic regression for indicator-based enrichment was used to calculate p values. Tiles labeled with −log10(P) and an asterisk represent statistically significant enrichment at FDR-adjusted p < 0.05. B and C: GO biological process pathway analyses (left) and temporal expression scatterplots (right) between PCW 9 and 34 months (M) for nsCS-enriched royalblue (B) and saddlebrown (C) modules. Red line indicates threshold p of 0.05. Asterisks represent significant differences in expression across modules at specific time points.
To investigate the developmental trajectories of the modules, we calculated the module eigengenes and correlated them with temporal time points (Fig. S7). The royalblue module is significantly expressed from PCW 9 to PCW 17 (p = 4 × 10−6) (Fig. 5B), while the saddlebrown module is significantly expressed at PCW 9 (p = 5 × 10−22) and PCW 16 (p = 5 × 10−6) (Fig. 5C). This is a critical epoch spanning the 1st and 2nd trimesters of human development and is correlated with formation and expansion of the cerebral cortex, including neuroprogenitor cell growth, neurogenesis, and neuronal differentiation.36 Consistent with this, GO enrichment analysis revealed that the royalblue module is enriched for terms related to epigenetic regulation of gene expression including histone modification, chromatin remodeling, H3, Set1C/COMPASS, MOZ/MORF, and MLL1/2 complexes (Fig. 5B, Fig. S8), the dysregulation of which is implicated in multiple neurodevelopmental disorders.37 The saddlebrown module is enriched for terms related to the formation of the cranial neural tube, including primary neural tube formation and tube closure (Fig. 5C, Fig. S8).
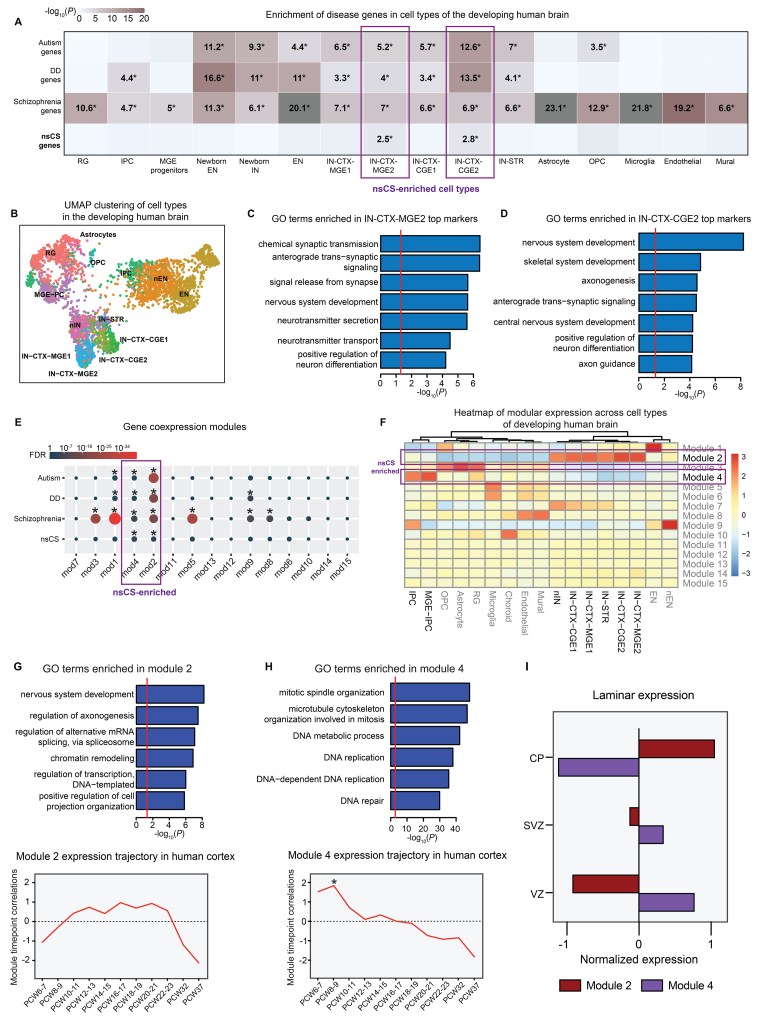
To determine the cell types during brain development that may be impacted by nsCS gene mutations, we studied the expression of nsCS-mutated genes in a single-cell transcriptomic atlas of human cortical development comprising 4261 cells across the human cortex spanning PCWs 5–37 (Fig. 6B).19 Enrichment was identified in two cell-type clusters: medial ganglionic eminence (MGE)–derived inhibitory neuron 2 (IN-CTX-MGE2; p = 3.1 × 10−3) and caudal ganglionic eminence (CGE)–derived inhibitory neuron 2 (IN-CTX-CGE2; p = 1.6 × 10−3) (Fig. 6A). Corroborating the overlap between nsCS and other neurodevelopmental disorders suggested by transcriptomic network analysis, nsCS-enriched inhibitory neuron populations are also enriched for autism, DD, and schizophrenia genes (Fig. 6A). In addition to enrichment of GO terms related to synaptic development such as anterograde transsynaptic signaling and neurotransmitter secretion, the top cell-type markers for nsCS-enriched inhibitory neuron clusters were also enriched for terms associated with corticogenesis (e.g., nervous system development, positive regulation of neuron differentiation) and suturogenesis (e.g., skeletal system development) (Fig. 6C and D, Fig. S9). The enrichment of nsCS and neuropsychiatric disorder risk genes in these cell types is consistent with previous work implicating altered development of fetal inhibitory neurons as a convergent cellular pathology in autism.38
FIG. 6.
Convergence of nsCS genes in gene coexpression networks and inhibitory neurons during human brain development in an scRNA-seq data set. A: Cell-type enrichment for disease risk genes. Tiles labeled with −log10(P) and an asterisk represent statistically significant enrichment at FDR-adjusted p < 0.05. B: UMAP clustering of developing human brain cell populations. C: GO biological process pathway analysis for top genes in nsCS-enriched IN-CTX-MGE2. Red line indicates threshold p value of 0.05. D: GO biological process pathway analysis for top genes in nsCS-enriched IN-CTX-CGE2. Red line indicates threshold p value of 0.05. E: Enrichment of disease risk genes in developing human brain gene coexpression modules. Analyzed transcriptomic data set from Nowakowski et al.19 The hypergeometric enrichment test was used to calculate p values. F: Heatmap of gene coexpression modules across individual cell types in developing human brain. G and H: GO biological process pathway analyses (upper) and temporal expression scatterplots (lower) between PCWs 5 and 37 for nsCS-enriched module 2 (G) and module 4 (H). Red line indicates threshold p of 0.05. Asterisks represent significant differences in expression across modules at specific time points. I: Laminar expression of gene coexpression networks in developing human brain. CP = cortical plate; EN = early- and late-born excitatory neurons; IN-STR = striatal neurons; MGE progenitors = MGE progenitor cells; mod = module; Mural = mural/pericytes; Newborn EN, nEN = newborn excitatory neurons; Newborn IN, nIN = newborn inhibitory neurons; OPC = oligodendrocyte progenitor cells; RG = radial glial cells; SVZ = subventricular zone; VZ = ventricular zone.
To better examine the functional convergence of nsCS-mutated genes during fetal brain development, we constructed gene coexpression networks from the single-cell transcriptomic atlas of human cortical development.19 We found module 2 (p = 4.9 × 10−3) and module 4 (p = 2.6 × 10−4) to be significantly enriched with nsCS-mutated genes (Fig. 6E). Both of these nsCS-enriched modules are also enriched for schizophrenia (module 2: p = 2.9 × 10−18; module 4: p = 9.0 × 10−7), autism (module 2: p = 1.2 × 10−28; module 4: p = 5.0 × 10−3), and DD (module 2: p = 4.9 × 10−20; module 4: p = 4.6 × 10−3) risk genes (Fig. 6E). At the cell-type level, module 4 is most highly expressed in intermediate progenitor cells (IPCs) and MGE-IPCs (Fig. 6F), which are IPCs derived from the cortex and MGE. Consistent with its cell-type expression profile, module 4 is most upregulated during PCWs 6–9 (Fig. 6H), an epoch crucial for the neural progenitor cell proliferation and differentiation necessary for normal brain development.
As expected, the top GO terms for module 4 include processes related to mitotic division such as DNA replication and mitotic spindle organization, while its hub genes consist of cell-cycle and proliferation genes (MKI67, TOP2A, CDK1, KIF11, KIF15, and KIF23) (Fig. 6H, Fig. S10). On the other hand, module 2 is most highly expressed in inhibitory neurons of MGE and CGE (Fig. 6F) in the cortical plate (Fig. 6I), where the module is upregulated around PCW 12 and remains elevated until PCWs 22–23 (Fig. 6G). Module 2’s hub genes include SOX transcription factors (SOX1, SOX2, SOX4, SOX6, and SOX11), which are crucial regulators of neurodifferentiation, and multiple chromatin remodeling genes (ARID1A, ARID1B, SATB1, SMARCC1, SMARCD1, SMARCE1, and BPTF), which are implicated in neurodevelopmental disorders.39 Module 2 genes are enriched for GO terms that implicate epigenetic (chromatin remodeling and regulation of transcription) and corticogenesis (nervous system development and regulation of axonogenesis) processes (Fig. 6G, Fig. S10).
Discussion
The genetic complexity of nsCS poses a major challenge in understanding disease mechanisms. In this study, we performed an integrative genomics analysis to help elucidate the biological pathways during mammalian neurocranial development in which nsCS gene mutations may converge to exert their pathogenic effects. We found that genes harboring DNMs predicted to be pathogenic in nsCS patients converged in cell types and gene coexpression networks within a developmental nexus of bone, meninges, and brain. We found that nsCS-enriched osteoprogenitor cells of the developing cranial suture do not act in isolation but appear rather to be part of an elaborate intercellular network inclusive of connective tissue cell types and neurons. We also found that nsCS-mutated genes converge in transcriptional modules implicating epigenetic regulation and neural tube formation during the first 2 trimesters of human brain development as well as fetal inhibitory neurons. Furthermore, nsCS-enriched transcriptomic networks and inhibitory neurons are enriched for autism and schizophrenia genes, implicating shared mechanisms responsible for abnormal cognition and neurobehavior in nsCS. These implicate the concurrent impact of nsCS-associated DNMs on cranial morphogenesis and brain development in mesenchymal osteogenic cells and parenchymal neural cells (see Fig. 7 for a summary of findings).
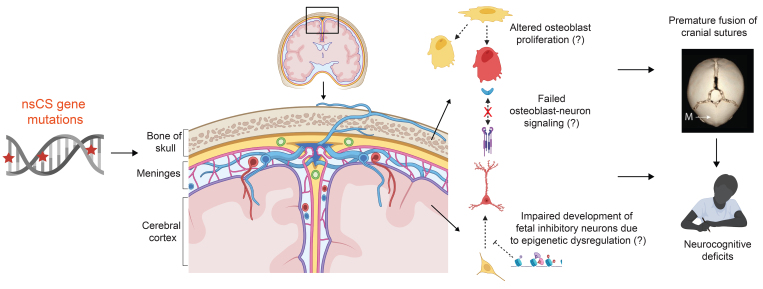
FIG. 7.
Impact of nsCS gene mutations on a developmental bone-meninges-brain nexus. CS gene mutations may influence neurodevelopment via multiple hypothesized direct and indirect mechanisms. First, nsCS gene mutations may disrupt the growth of suture osteoblasts, which do not act in isolation but are part of intercellular signaling networks that include other cell types in the meninges and brain. Thus, abnormal development of suture osteoblasts not only contributes to premature fusion of cranial sutures, but also affects development of the brain parenchyma due to altered mesenchymal-neuronal signaling crosstalk. Second, CS gene mutations directly impact brain development by influencing the maturation of fetal inhibitory neurons. The convergence of CS genetic risk on a developmental bone-meninges-brain nexus therefore explains the pathogenesis of bone deformity underlying premature fusion of cranial sutures as well as neuronal maldevelopment that underlies the persistence of neurocognitive deficits despite optimal surgical management. This figure was created with Biorender.com.
A clinical conundrum in nsCS is why affected children continue to exhibit subtle neurocognitive deficits despite optimal surgical management of the skull deformity.6,7 Nonsyndromic CS is classically considered a primary bone disorder that exerts secondary neurological injury by the mechanical hindrance of brain expansion from poor compliance of the affected cranial vault. Thus, functional investigations have focused on the impact of nsCS-associated genes on mesenchymal tissue development such as the proliferation and growth of cranial suture osteoprogenitors.8,12,16 Our functional genomics analyses here show that nsCS-associated DNMs may have an impact on brain development at the cellular and molecular levels. We show that osteoprogenitors communicate with neurons via the BMP, FGF, and ncWNT pathways as part of intercellular signaling networks. Perturbation of osteoprogenitors may therefore affect cerebral cortex development by disrupting this mesenchymal-neural signaling axis (Fig. 7).
The in silico findings presented here are corroborated by functional studies showing that cranial connective tissues are not mere supportive bystanders but rather have active roles in shaping brain development.40 A particularly striking example is the finding that loss of meninges-specific deletion of Twist1, a known syndromic nsCS gene, alters the formation of the cerebral cortex.40 Second, we identify inhibitory neurons in the MGE and CGE as cell types in the developing human brain that are enriched for nsCS-mutated genes, suggesting an impact of nsCS genetic risk on the brain parenchyma itself. These findings implicate mesenchymal-neural signaling and fetal inhibitory neurons as developmental processes and cell types, respectively, that may be studied in future functional studies to clarify the etiology of neurocognitive deficits in children with nsCS.
Surgical repair of the cranial defect is the primary treatment strategy for nsCS.41 However, there remains controversy regarding the timing and type of surgical intervention, namely open vault repair versus endoscopic strip craniectomy.42 Clinically speaking, our findings suggest an important role of genetics in neurodevelopment, suggesting that long-term outcomes may not be driven by the specific surgical approach, but rather by whether the child harbors a pathogenic gene mutation.11,43,44 This corroborates and extends previous clinical-genetic work that has shown that a patient’s genotype predicts neurodevelopmental outcomes in children with nsCS, such as the presence of a mutation in SMAD6.43,44 While further investigation is still needed to clarify the appropriate timing and surgical intervention type, the impact of genetics on the neurodevelopmental pathology of nsCS may warrant exome sequencing as a diagnostic adjunct in the clinical evaluation of nsCS children. In the long term, a more complete understanding of the molecular genetic mechanisms underlying nsCS will increase precision in the diagnosis, prognosis, and treatment of patients.
There are limitations to our study. While our in silico analysis demonstrates several genetic mechanisms for nsCS pathophysiology, further investigation is needed for functional validation of our findings via additional experimental designs. Addressing this will strengthen the validity and real-world applicability of our results, contributing to a more comprehensive understanding of nsCS pathophysiology.
Conclusions
Our findings indicate that nsCS-associated DNMs have a simultaneous effect on cranial morphogenesis and cortical development through both cell- and non–cell-autonomous mechanisms involving fetal osteoblasts, pial fibroblasts, and neurons in a developmental context. These results suggest that the mutational status might have a greater influence on neurodevelopmental outcomes in nsCS patients than the surgical technique used.
Acknowledgments
This work was funded by the National Institute of Neurological Disorders and Stroke (grant no. RO1NS111029 to K.T.K.), National Institute of General Medical Sciences (grant no. T32GM136651 to P.Q.D.), and Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant no. F30HD106694 to P.Q.D.).
Disclosures
Dr. Alper reported consulting fees from Decibel Therapeutics, D.E. Shaw Research, and Entrada Therapeutics; and research support from Quest Diagnostics, Inc.
Author Contributions
Conception and design: Kahle, Kiziltug, Duy, Allington, Timberlake, DiLuna, Alperovich. Acquisition of data: Kahle, Kiziltug, Allington, Timberlake, Long, DiLuna. Analysis and interpretation of data: Kahle, Kiziltug, Duy, Allington, Timberlake, Kawaguchi, Alper, Alperovich, Geschwind. Drafting the article: Kahle, Kiziltug, Duy, Allington, DiLuna. Critically revising the article: Kahle, Kiziltug, Duy, Allington, Long, Almeida, DiLuna, Alper, Alperovich, Geschwind. Reviewed submitted version of manuscript: Kahle, Kiziltug, Duy, Allington, Timberlake, Kawaguchi, Almeida, DiLuna, Alper, Alperovich. Approved the final version of the manuscript on behalf of all authors: Kahle. Statistical analysis: Kiziltug, Allington, Timberlake, Kawaguchi. Administrative/technical/material support: Kahle, DiLuna. Study supervision: Kahle, Kiziltug.
Supplemental Information
- Tables S1–S5 and Figs. S1–S10. https://thejns.org/doi/suppl/10.3171/2023.8.PEDS23155.
References
- 1. Richtsmeier JT, Flaherty K. Hand in glove: brain and skull in development and dysmorphogenesis. Acta Neuropathol. 2013;125(4):469–489. doi: 10.1007/s00401-013-1104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegenthaler JA, Pleasure SJ. We have got you ‘covered’: how the meninges control brain development. Curr Opin Genet Dev. 2011;21(3):249–255. doi: 10.1016/j.gde.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boulet SL, Rasmussen SA, Honein MA. A population-based study of craniosynostosis in metropolitan Atlanta, 1989-2003. Am J Med Genet A. 2008;146A(8):984–991. doi: 10.1002/ajmg.a.32208. [DOI] [PubMed] [Google Scholar]
- 4. Lajeunie E, Le Merrer M, Bonaïti-Pellie C, Marchac D, Renier D. Genetic study of nonsyndromic coronal craniosynostosis. Am J Med Genet. 1995;55(4):500–504. doi: 10.1002/ajmg.1320550422. [DOI] [PubMed] [Google Scholar]
- 5. Persing JA. MOC-PS(SM) CME article: management considerations in the treatment of craniosynostosis. Plast Reconstr Surg. 2008;121(4 suppl):1–11. doi: 10.1097/01.prs.0000305929.40363.bf. [DOI] [PubMed] [Google Scholar]
- 6. Kapp-Simon KA, Figueroa A, Jocher CA, Schafer M. Longitudinal assessment of mental development in infants with nonsyndromic craniosynostosis with and without cranial release and reconstruction. Plast Reconstr Surg. 1993;92(5):831–841. [PubMed] [Google Scholar]
- 7. Chieffo D, Tamburrini G, Massimi L, et al. Long-term neuropsychological development in single-suture craniosynostosis treated early. J Neurosurg Pediatr. 2010;5(3):232–237. doi: 10.3171/2009.10.PEDS09231. [DOI] [PubMed] [Google Scholar]
- 8. Twigg SRF, Wilkie AOM. A genetic-pathophysiological framework for craniosynostosis. Am J Hum Genet. 2015;97(3):359–377. doi: 10.1016/j.ajhg.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Timberlake AT, Kiziltug E, Jin SC, et al. De novo mutations in the BMP signaling pathway in lambdoid craniosynostosis. Hum Genet. 2023;142(1):21–32. doi: 10.1007/s00439-022-02477-2. [DOI] [PubMed] [Google Scholar]
- 10. Timberlake AT, Furey CG, Choi J, et al. De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proc Natl Acad Sci U S A. 2017;114(35):E7341–E7347. doi: 10.1073/pnas.1709255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duy PQ, Timberlake AT, Lifton RP, Kahle KT. Molecular genetics of human developmental neurocranial anomalies: towards "precision surgery.". Cereb Cortex. 2023;33(6):2912–2918. doi: 10.1093/cercor/bhac249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teng CS, Ting MC, Farmer DT, Brockop M, Maxson RE, Crump JG. Altered bone growth dynamics prefigure craniosynostosis in a zebrafish model of Saethre-Chotzen syndrome. eLife. 2018;7:e37024. doi: 10.7554/eLife.37024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parikshak NN, Luo R, Zhang A, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Willsey AJ, Sanders SJ, Li M, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duy PQ, Weise SC, Marini C, et al. Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus. Nat Neurosci. 2022;25(4):458–473. doi: 10.1038/s41593-022-01043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farmer DT, Mlcochova H, Zhou Y, et al. The developing mouse coronal suture at single-cell resolution. Nat Commun. 2021;12(1):4797. doi: 10.1038/s41467-021-24917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeSisto J, O’Rourke R, Jones HE, et al. Single-cell transcriptomic analyses of the developing meninges reveal meningeal fibroblast diversity and function. Dev Cell. 2020;54(1):43–59.e4. doi: 10.1016/j.devcel.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M, Santpere G, Imamura Kawasawa Y, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362(6420):eaat7615. doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowakowski TJ, Bhaduri A, Pollen AA, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358(6368):1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruzzo EK, Pérez-Cano L, Jung JY, et al. Inherited and de novo genetic risk for autism impacts shared networks. Cell. 2019;178(4):850–866.e26. doi: 10.1016/j.cell.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satterstrom FK, Kosmicki JA, Wang J, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180(3):568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McRae JF, Clayton S, Fitzgerald TW, et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Federico A, Monti S. hypeR: an R package for geneset enrichment workflows. Bioinformatics. 2020;36(4):1307–1308. doi: 10.1093/bioinformatics/btz700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(Article17) doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 30. Walker RL, Ramaswami G, Hartl C, et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell. 2019;179(3):750–771.e22. doi: 10.1016/j.cell.2019.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Timberlake AT, McGee S, Allington G, et al. De novo variants implicate chromatin modification, transcriptional regulation, and retinoic acid signaling in syndromic craniosynostosis. Am J Hum Genet. 2023;110(5):846–862. doi: 10.1016/j.ajhg.2023.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Como CN, Kim S, Siegenthaler J. Stuck on you: meninges cellular crosstalk in development. Curr Opin Neurobiol. 2023;79:102676. doi: 10.1016/j.conb.2023.102676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Selvaraj P, Huang JS, Chen A, Skalka N, Rosin-Arbesfeld R, Loh YP. Neurotrophic factor-α1 modulates NGF-induced neurite outgrowth through interaction with Wnt-3a and Wnt-5a in PC12 cells and cortical neurons. Mol Cell Neurosci. 2015;68:222–233. doi: 10.1016/j.mcn.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 35. Tillman KK, Höijer J, Ramklint M, Ekselius L, Nowinski D, Papadopoulos FC. Nonsyndromic craniosynostosis is associated with increased risk for psychiatric disorders. Plast Reconstr Surg. 2020;146(2):355–365. doi: 10.1097/PRS.0000000000007009. [DOI] [PubMed] [Google Scholar]
- 36. Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89(2):248–268. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paulsen B, Velasco S, Kedaigle AJ, et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602(7896):268–273. doi: 10.1038/s41586-021-04358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsurusaki Y, Okamoto N, Ohashi H, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44(4):376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 40. Ho-Nguyen K, Jain M, Matrongolo MJ, Ang PS, Schaper S, Tischfield MA. Twist1 and balanced retinoic acid signaling act to suppress cortical folding in mice. bioRxiv. doi: 10.1101/2022.09.27.509818. Preprint posted online November 15, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marbate T, Kedia S, Gupta DK. Evaluation and management of nonsyndromic craniosynostosis. J Pediatr Neurosci. 2022;17(suppl 1):S77–S91. doi: 10.4103/jpn.JPN_17_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shah MN, Kane AA, Petersen JD, Woo AS, Naidoo SD, Smyth MD. Endoscopically assisted versus open repair of sagittal craniosynostosis: the St. Louis Children’s Hospital experience. J Neurosurg Pediatr. 2011;8(2):165–170. doi: 10.3171/2011.5.PEDS1128. [DOI] [PubMed] [Google Scholar]
- 43. Timberlake AT, Junn A, Flores R, Staffenberg DA, Lifton RP, Persing JA. Genetic influence on neurodevelopment in nonsyndromic craniosynostosis. Plast Reconstr Surg. 2022;149(5):1157–1165. doi: 10.1097/PRS.0000000000008976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu RT, Timberlake AT, Abraham PF, et al. SMAD6 genotype predicts neurodevelopment in nonsyndromic craniosynostosis. Plast Reconstr Surg. 2020;145(1):117e–125e. doi: 10.1097/PRS.0000000000006319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- Tables S1–S5 and Figs. S1–S10. https://thejns.org/doi/suppl/10.3171/2023.8.PEDS23155.