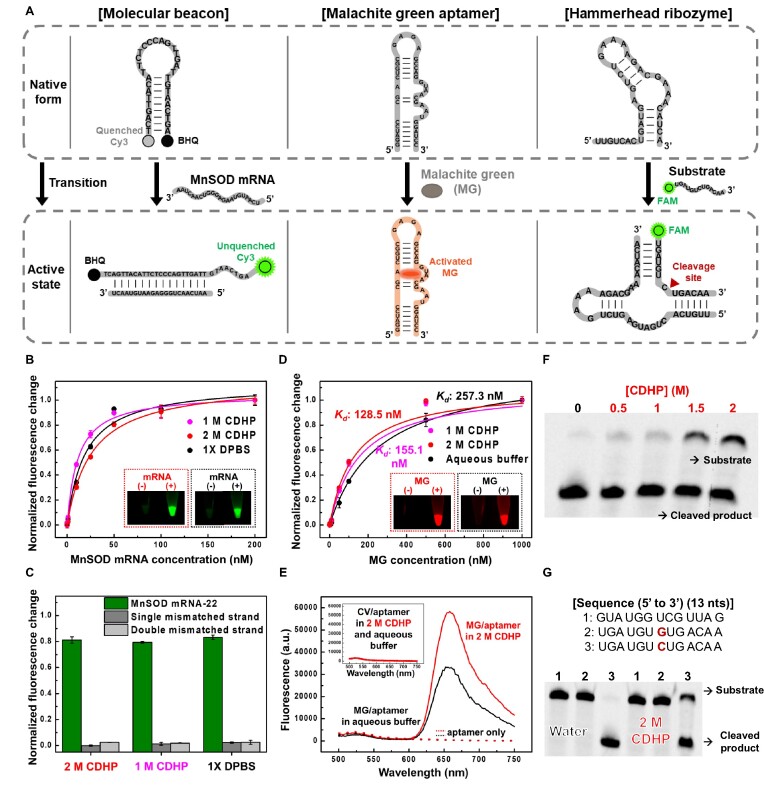
Figure 2.
Confirmation of active FNAs in the CDHP cage. (A) Three different FNAs (MnSOD mRNA-sensing MB, MG aptamer and hammerhead ribozyme) and their pre-defined molecular functions. In response to cognate targets or substrates, the FNAs can be activated to perform specific target recognition or substrate cleavage, synchronized with detectable fluorescent changes. (B) The MnSOD MB-based mRNA detection in CDHP (1 and 2 M) and 1× DPBS. Like 1× DPBS, the CDHP allowed RNA:DNA hybridization to induce a large conformational change of MnSOD MB in emitting a strong fluorescent signal. As a result, we observed similar signaling patterns in a target concentration-dependent manner. (C) Conserved target specificity of MnSOD MB. Either in CDHP or in 1× DPBS, the MnSOD MB could discriminate its target RNA from single or double mismatched non-target RNAs. (D) Aptameric MG recognition and fluorescent increases in CDHP (1 and 2 M) and aqueous buffer. In the CDHP, the MG-specific aptamer could recognize its target fluorogen to perform binding-induced fluorescent enhancing, even exhibiting lower Kds than in the aqueous buffer. (E) Conserved target specificity of MG-specific aptamer. Unlike MG, its structural analog, CV, could not be bound to the MG-specific aptamer without fluorogen stabilization, leading to no increase in fluorescence either in CDHP or in aqueous buffer. (F) Ribozyme-mediated substrate cleavage at different CDHP concentrations. The hammerhead ribozyme could perform the desired phosphoryl transfer reaction in the CDHP with slightly lower yields than in water. (G) Conserved substrate specificity of hammerhead ribozyme. The ribozyme selectively cleaved its target substrate both in water and 2 M CDHP.