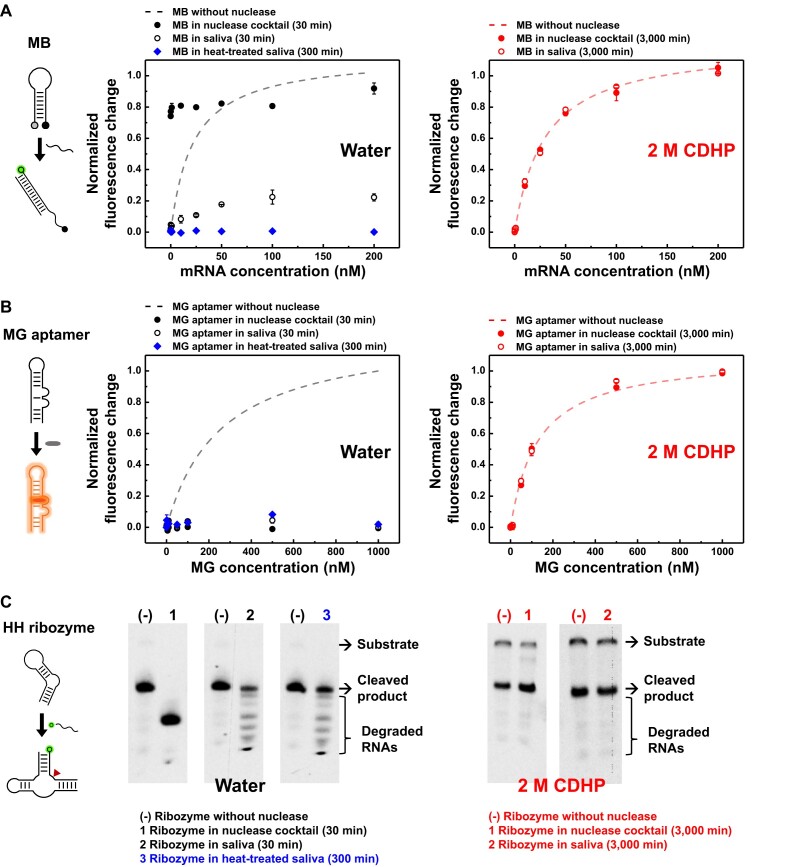
Figure 5.
CDHP-mediated shielding of active FNAs in nuclease cocktails and human saliva. (A) MB-based mRNA detection with or without 2 M CDHP. Both nuclease cocktails and saliva (non- or heat-treated) permitted nucleolytic digestion of MnSOD MB or MnSOD mRNA-22 without CDHP, relevant to false-positive or true-negative detection errors, respectively (left). With 2 M CDHP, however, both MnSOD MB and MnSOD mRNA-22 were fully shielded even in the presence of undesired and unknown nucleases during 3000 min incubation at 37°C, observing desired fluorescent signals in a target concentration-dependent manner like the intact MB with no nuclease (right). (B) Target recognition of MG-specific aptamers with or without 2 M CDHP. Without CDHP, high RNase activities in nuclease cocktails and saliva (non- or heat-treated) caused full hydrolysis of MG-specific RNA aptamers, and no aptamer-bound stabilization of MGs was detected with observation of true-negative fluorescent signals (left). However, use of 2 M CDHP effectively inhibited the RNase activities over 3000 min at 37°C, allowing the MG-specific aptamer to perform binding-induced fluorescent enhancing in a MG concentration-dependent manner with no decrease in affinity (right). (C) Ribozyme-mediated substrate cleavage with or without 2 M CDHP. The absence of CDHP revealed the undesirable loss of nucleic acids, including cleaved products, both in nuclease cocktails and human saliva (non- and heat-treated) (left). On the other hand, the CDHP cage allowed desired substrate-specific cleavage reactions irrespective of the presence of various nucleases, and both uncleaved substrates and cleaved products were well conserved even after 3000 min incubation at 37°C (right). For all the experiments, the heat treatment of saliva was performed at 90°C for 15 min.