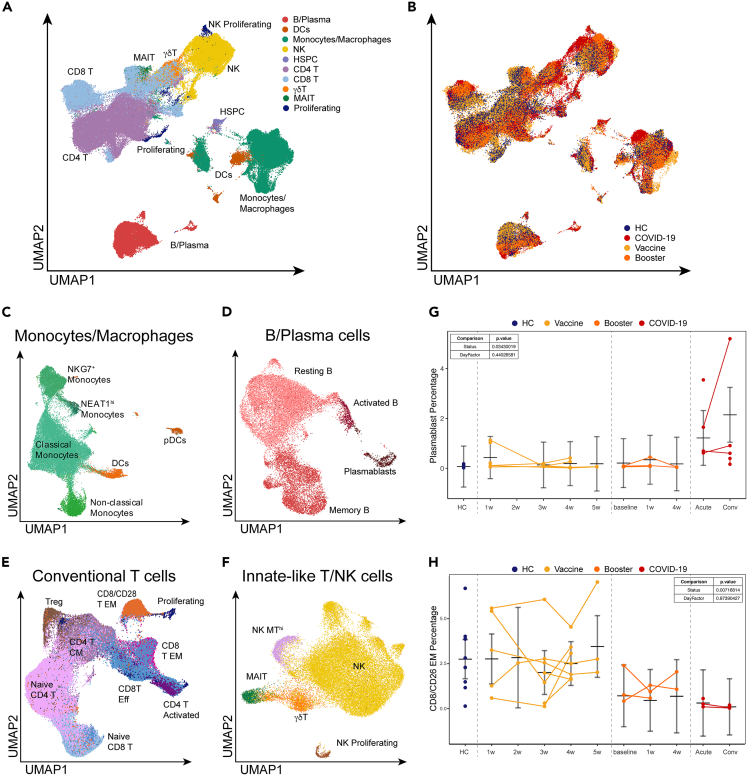
Figure 1.
Single-cell landscape of immunological responses to COVID-19 and SARS-CoV-2 BNT162b2 mRNA vaccine
(A) UMAP representation of over 195,000 PBMCs by scRNA-seq, clustered and colored by. indicated cell type. Clusters identified based on gene expression and surface epitopes.
(B) UMAP visualization of PBMCs from and COVID-19-naïve healthy donors (blue), healthy volunteers before receiving the BNT162b2 mRNA vaccine (blue), healthy volunteers after receiving the BNT162b2 mRNA vaccine (yellow) and booster (orange), and COVID-19 patients (red).
(C–F) UMAP representation of subclustered myeloid (C), B cell (D), T cell (E), and innate and unconventional T cell (F) populations colored and labeled by cell type.
(G and H) Graphs highlight specific populations that exhibited significant differences between healthy volunteers and COVID-19 patients. Vaccine samples are shown as weeks after first vaccine dose. Booster samples are shown as weeks after booster. COVID-19 patient samples are split by days post-onset (DPO) of symptoms into acute (≤10 DPO) and convalescent (>10 DPO). Line graphs show percentages for given cell population per subject, per time point. Connected lines indicate repeated measurements for the same subjects. P-values for these plots are determined by an ANOVA of linear mixed models and post-hoc pairwise comparison of estimated marginal means are listed on each plot. The bars represent the upper and lower confidence intervals for the estimated marginal means for the linear model.