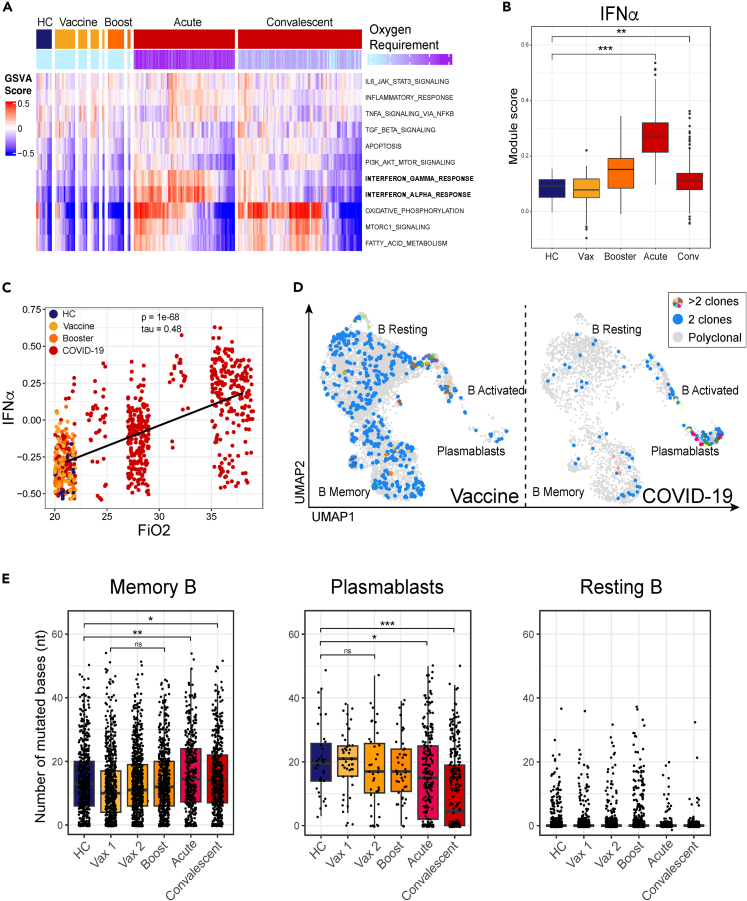
Figure 2.
Maturation of B cell responses
(A) GSVA analysis of plasmablasts in cells from HC, Vaccine and COVID-19 (top color bar) patients from the Hallmark gene set32 and colored by oxygen requirement (bottom color bar) which is a clinical parameter defined as the fraction of inspired oxygen, where 21% represents oxygen content of room air without supplementation. HC, Vaccine, and Booster cells are colored at 21%. COVID-19 patient samples are split into acute (≤10 DPO) and convalescent (>10 DPO). See also Figure S4.
(B) IFN-I gene pathway module score in plasmablasts. Module scores were calculated using the AddModuleScore function from the Seurat package, which calculates the mean expression for a set of genes and adjusts for the collective expression of control features. Box and whisker plots were used to portray the distribution of the data, outliers, and the median. P-values were determined by the Wilcoxon test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). See also Figures S4 and S5.
(C) Oxygen requirement (shown as Fraction of inspired oxygen, FiO2) against GSVA enrichment score for interferon alpha response gene set as shown in A. P and tau values are determined by a Kendall rank correlation test.
(D) Clonal populations based on individual B cell IGH chain CDR3 sequence. CDR3 sequences occurring in at least 2 cells are colored blue, while any CDR3 sequences in at least 3 cells are colored uniquely in cells from Vaccinated (left) and COVID-19 patient samples (right).
(E) Number of mismatched bases according to IgBlast results of recovered VH gene sequences in memory (left), plasmablasts (middle), and resting (right) B cell subsets in cells from HC (blue), Vaccinated (yellow), Boosted (orange), and COVID-19 (red) patient samples. Box and whisker plots were used to portray the distribution of the data, outliers, and the median. P-values were determined by Wilcoxon test (ns p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).