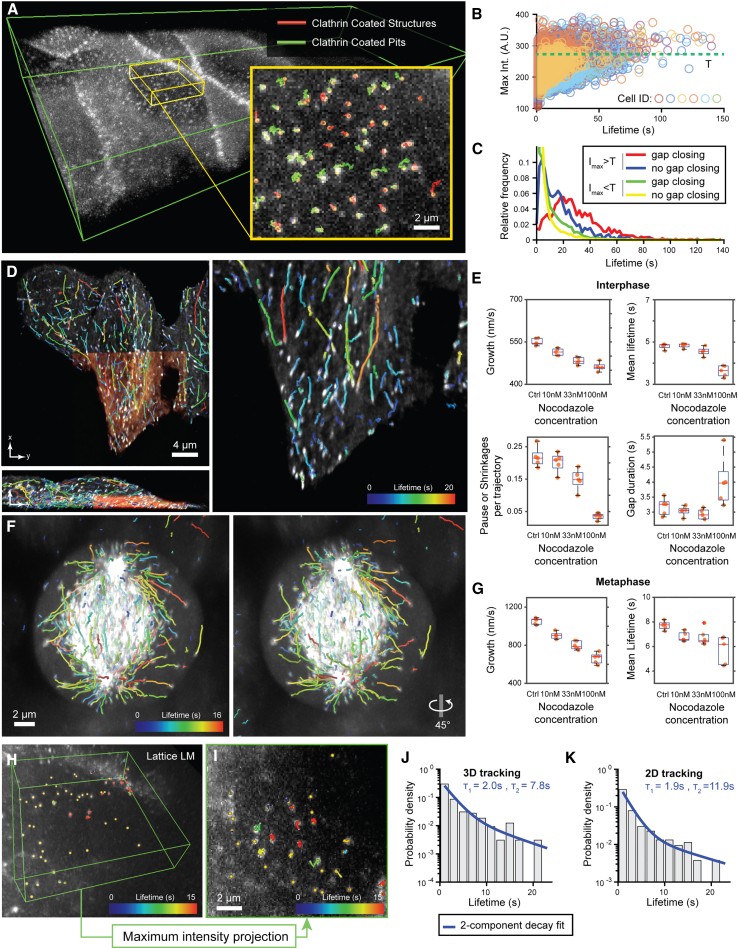
Figure 2.
u-track3D supports a variety of imaging and biological scenarios
(A) Maximum intensity projections (MIPs) of a rat kidney cell layer imaged with diagonally scanned light-sheet microscopy (diaSLM). Cells are expressing eGFP-labeled alpha subunit of the AP-2 complex. Green box is 160 × 40 × 12 μm. Inset shows trajectories of clathrin aggregates classified as clathrin-coated structures or maturing pits.
(B) Normalized maximum intensity of each trajectory as a function of lifetime plotted for six cellular layers composed of multiple cells each. The green line denotes the median of the cumulated distribution (value T).
(C) Probability density of lifetime for the set of trajectories above and below the threshold value T, with and without gap closing (n = 6 cellular layers, pooled trajectories lifetimes).
(D) MIP of HeLa cells in interphase imaged with lattice light-sheet microscopy (LLSM) expressing eGFP-labeled EB1 (orange area is 30 × 32 × 7 μm). Overlay highlights EB1 trajectories.
(E) Average microtubule lifetimes, microtubule growth rate, as well as average number and duration of pause and shrinkage events per trajectory for increasing concentrations of nocodazole (n = 5 per conditions; center line, median; box limits, 25 and 75 percentiles; whiskers, extremum).
(F) MIP of HeLa cells in metaphase imaged with LLSM along with 45°rotation around the vertical axis. Overlay highlights EB1 trajectories.
(G) Same as (E) measured for cells in metaphase (n = 5 per conditions).
(H) MIP of mouse embryonic stem (ES) cell nucleus imaged with LLSM expressing GFP-labeled TFs. Green box is 13 × 13 × 3 μm. Overlay highlights SOX2 trajectories. (I) MIP of ES cell nucleus imaged with LLSM expressing GFP-labeled TFs. Overlay highlights SOX2 trajectories tracked after MIP transformation.
(J) Probability density of SOX2 binding time measured in LLSM overlaid with a two-component decay fit (n = 1 cell).
(K) Probability density of SOX2 binding time measured in projected LLSM data overlaid with a two-component decay fit (n = 1 cell).