Figure 5.
DynROIs drive the visualization of chromosome capture by microtubules and reveal possible interactions between neighboring kinetochore fibers
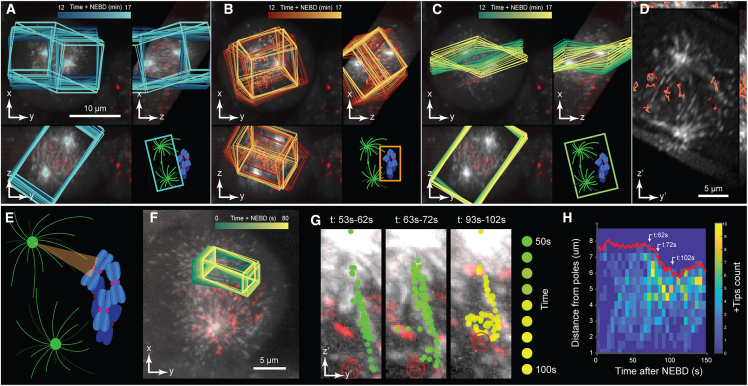
(A–D) Dual-colored orthogonal MIP of HeLa cells undergoing mitosis labeled with eGFP-labeled EB3 (marking microtubule plus-ends rendered in gray) and mCherry-labeled centromere protein A (marking kinetochores rendered in red). Overlays highlight (A) a dynROI built around centrosome trajectories, (B) a dynROI built around kinetochores trajectories, and (C) a plane built to visualize the dynamics of chromosomes relative to the spindle location. (D) View of the dynROI following description in (H).
(E) Definition of a conical dynROI between a centrosome and a kinetochore.
(F) Dual-colored orthogonal MIP of HeLa cells during prometaphase. Overlay highlights the motion of the dynROI.
(G) Cumulative overlays of the detected microtubule plus-end position for three periods of 10 s between 53 and 102 s post nucleus envelope breakage.
(H) Plus-ends count function of time and distance from the pole (n = 1 dynROI).