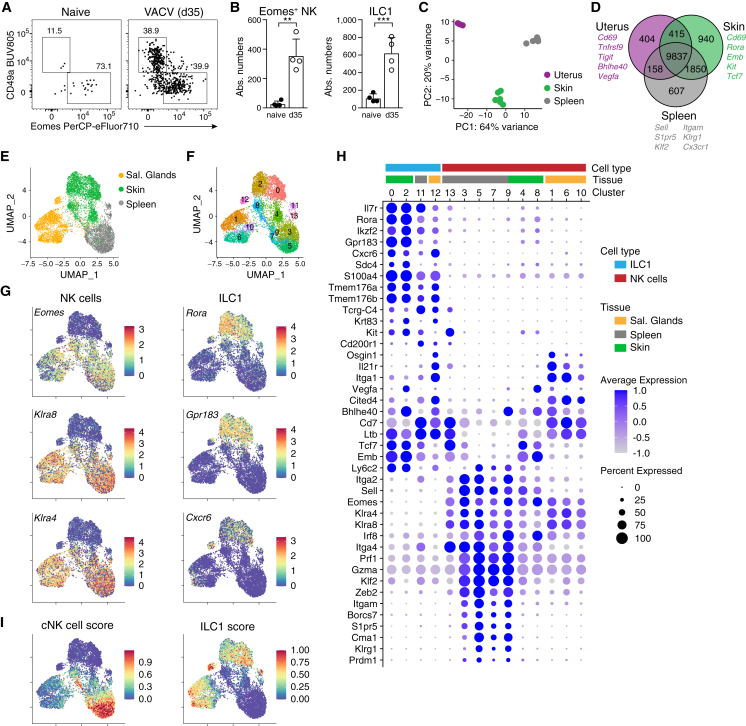
Figure 2.
Infection induces NK cells with a program of tissue-resident cells
(A and B) Representative FACS plots (A) and numbers of Lin−NK1.1+ pregated CD49a−Eomes+ NK cells and CD49a+Eomes− ILC1s (B) in naive versus VACV-infected ears on day 35 pi.
(C and D) mRNA-sequencing analysis of Lin−NK1.1+Cxcr6− NK cells sorted from naive (uterus) or VACV-infected mice on day 35 pi (spleen and skin). (C) Euclidean distance of samples visualized by principal-component analysis. (D) Venn diagram showing numbers and examples of shared and differentially expressed genes.
(E–I) scRNA-seq analysis of Lin−NK1.1+ cells sorted on day 25 post VACV infection from indicated organs, pooled from n = 5–10 individual mice each. UMAP visualization of tissue of origin (E), identified clusters (F), selected marker genes delineating NK cells and ILC1s (G), and scores based on the top 100 DEGs between NK cells and ILC1s (I).
(H) Bubble plot representation of Z score of selected marker gene expression.
Data are representative of three (A and B) or two (E–I) independent experiments with n = 4–18 ears per group. Error bars indicate mean + SD. p values were calculated by unpaired Student’s t test. ∗∗p < 0.01 and ∗∗∗p < 0.001. See also Figure S2.