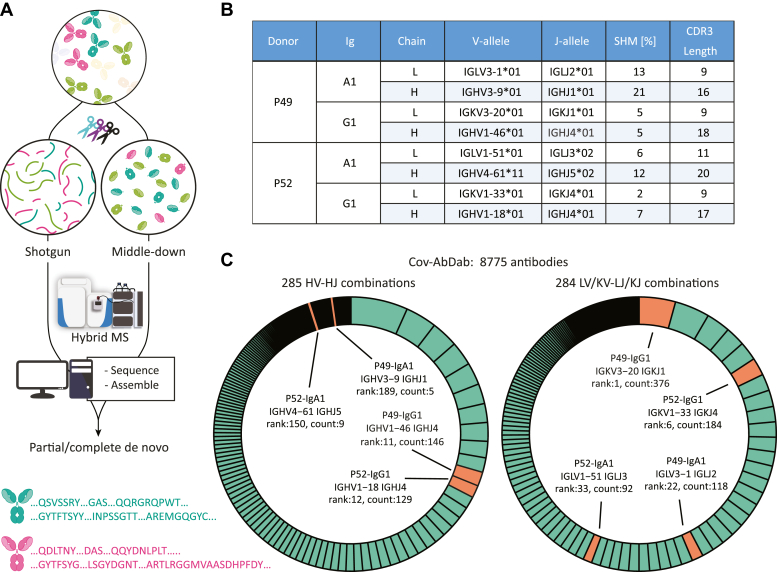
Fig. 5.
De novo sequencing combining peptide- and protein-centric approaches reveals that the dominant clones in P49 and P52 exhibit sequence features alike those deposited in the Cov-AbDab database of antibodies that bind and/or neutralize SARS-CoV-2.A, abundant IgA1 and IgG1 clones were fully de novo sequenced using an integrative MS approach, combining peptide-centric (shotgun) and protein-centric (middle-down) approach. In the peptide-centric approach, the Fab molecules were digested in parallel with a mix of proteases to provide a set of large set overlapping peptides covering the sequence of the Fab. In the protein-centric MS approach, the Fab molecules mass-selected and fragmented with electron-transfer dissociation providing sequence tags and mass-restraints that were used to score and refine the sequences predicted from the bottom-up approach. The protein-centric approach was performed both at the intact Fab-level and following reduction of the disulfide bridges in the Fab, by mass-selecting ions of either the Fab light-chain or Fd heavy chains. B, overview of the best matching germline alleles, the amount of somatic hypermutations and the CDR3 lengths for the fully sequenced most dominant IgG1 and IgA1 clones from the serum of P49 and P52. C, assignment (depicted in orange) of the IGHV-IGHJ and IG[K/L]V-IG[K/L]J gene pairs from the here determined clones (see 5B) among the unique pairs in the database of SARS-CoV-2-related antibodies (8775 total entries in Cov-AbDab, with categories depicted in green in the donut charts). The antibodies in the Cov-AbDab database are primarily discovered by B cell sequencing. Especially, the germline sequences from the IgG1 clones identified in this study match well within some of the most copious categories in the Cov-AbDab database. CDR3, complementarity determining region 3; Cov-AbDab, COVID antibody database; SARS-CoV-2, severe acute respiratory syndrome corona virus 2.