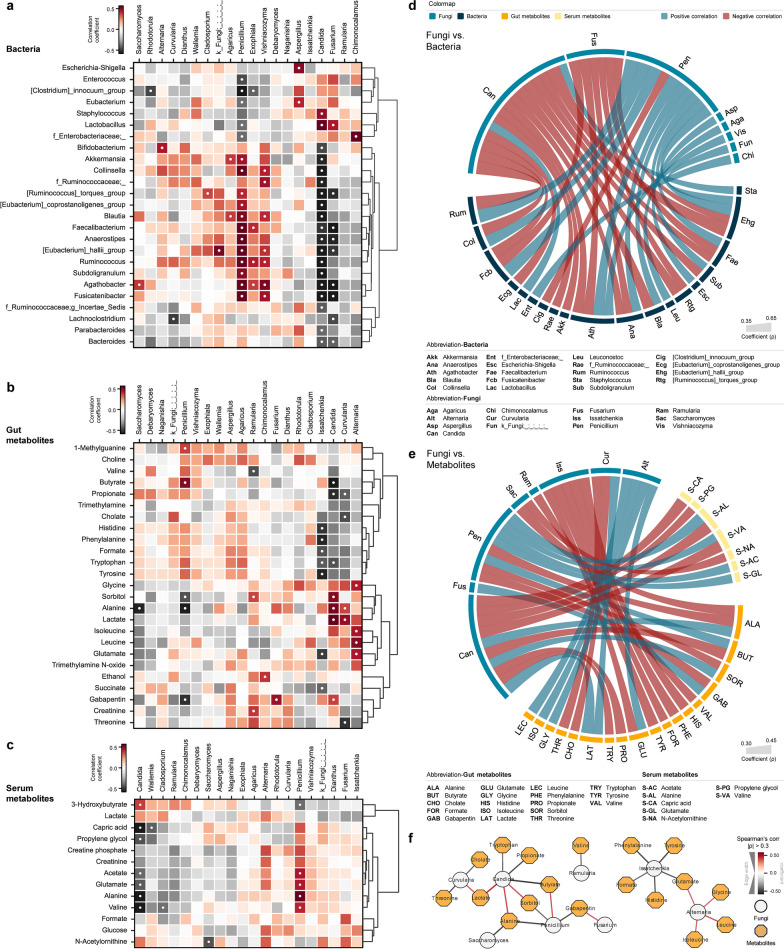
Fig. 3.
Mycobiome dysbiosis signatures correlate with perturbed gut bacteriome and gut-plasma metabolome arrays. Correlation arrays between mycobiome and a microbiome, b gut metabolites, and c plasma metabolites calculated with all samples from control, trauma and sepsis cohorts (Statistical significance: ● p < 0.05) Significant correlation networks d between mycobiome and microbiome (shown ρ > 0.35) and e between mycobiome and gut and plasma metabolites (shown ρ > 0.35). f Mycobiome-gut metabolites co-occurrence network. Circular nodes represent Fungi; while, yellow octagon nodes represent gut metabolites. Only significant links are shown here (Spearman’s rank correlation coefficient (ρ) > 0.3; Benjamini–Hochberg corrected p < 0.05). Red links denote positive correlation and black links indicate negative correlation, with line thickness corresponding to the correlation coefficient value