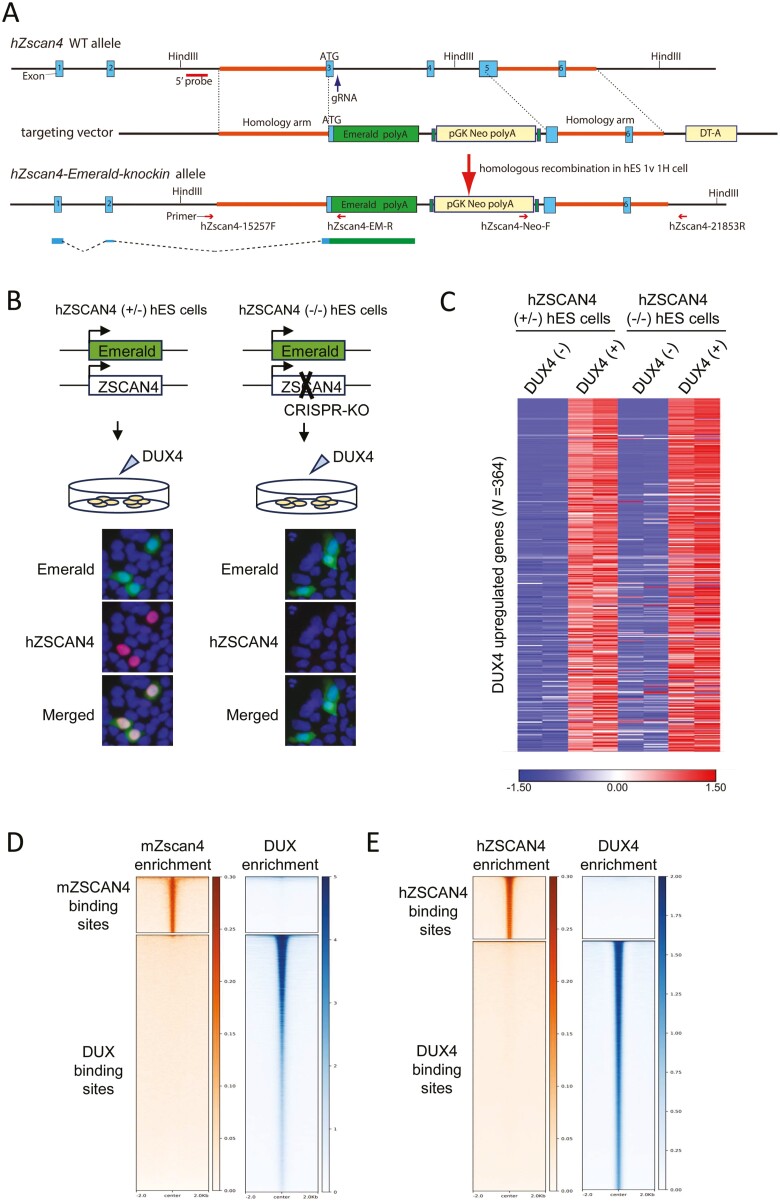
Figure 6.
hZSCAN4 deletion does not affect gene expression changes induced by DUX4. (A) Schematic illustrations of the hZSCAN4 wildtype allele, the ZSCAN4-Emerald knock-in allele and the targeting vector. Coding exon 3 is followed by Emerald and polyA signal. 5' probe for Southern blotting is shown. (B) Experimental scheme for generating hZSCAN4 (+/−) and (−/−) hES cells, in which Emerald GFP replaces one allele of hZSCAN4, and the other allele is either intact (+/−) or disrupted by CRISPR-Cas9 (−/−). Those cells were transfected with synthetic mRNA of DUX4 to induce DUX4-regulated transcriptional burst, including hZSCAN4 expression. Immunostaining confirmed that hZSCAN4 was expressed in hZSCAN4 (+/−) cells but not in hZSCAN4 (−/−) cells. Emerald GFP was expressed in both cell lines. Nuclei were stained with DAPI (Blue). (C) RNA-sequencing analysis showed that transcriptional burst was similarly detected in hZSCAN4 (+/−) cells and hZSCAN4 (−/−) cells. Three hundred and sixty-four genes are upregulated by DUX4 induction in hZSCAN4 (+/−) cells (fold change > 2 compared with non-transfected cells). The data from two biological replicates are shown. The colour scale bar shows z-score values. (D) Density heatmaps showing enrichment levels of mZSCAN4 (this study) and DUX (GSE95517_mDUX-HA-rep1.bw)12 within a 4 kb window centred at mZSCAN4 and DUX binding sites. (E) Density heatmaps showing enrichment levels of hZSCAN4 (this study) and DUX4 (GSM2515762_DUX4-rep2.bw)12 within a 4 kb window centred at hZSCAN4 and DUX4 binding sites.