Fig. 5 |. Condensate mediated transcriptional amplification.
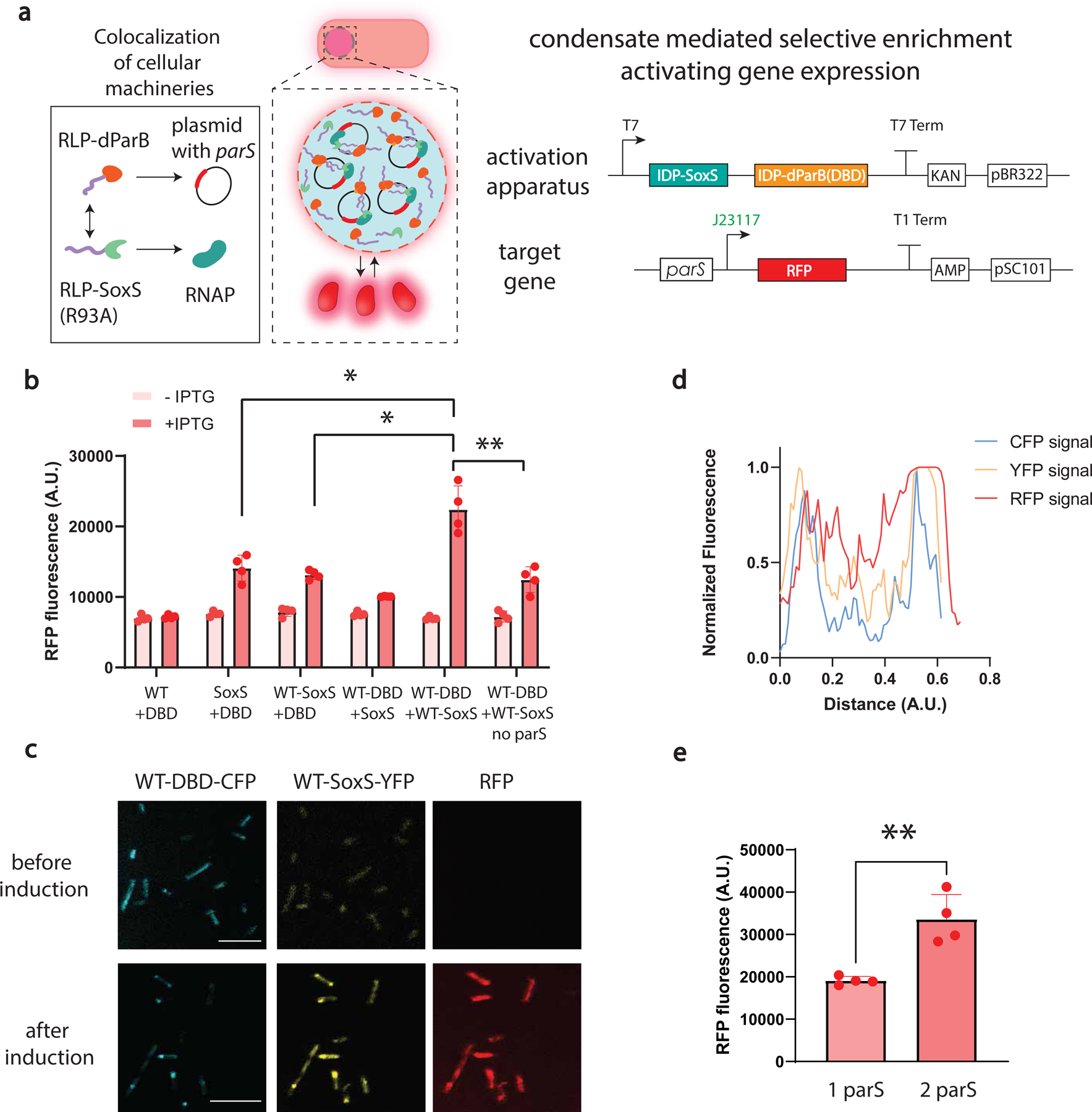
a, A dynamic condensate clusters key transcriptional machinery and plasmids, amplifying transcription because of the increased local concentrations of transcriptional activator and RNA polymerase in the condensate. Synthetic transcriptional machinery is constructed by synIDP-DBD directed condensate formation on parS sites and synIDP-SoxS (R93A) mediated recruitments of RNA polymerase to a weak promoter J23117. The synIDP-mediated condensate formation activates expression of the target gene. Expression of synIDP-dParB and synIDP-SoxS (R93A) is controlled by a T7 promoter.
b, Comparison of RFP fluorescence signal based on the combinations of different components, DBD, synIDP, SoxS (R93A) after 2h of induction of the components of the transcription activation apparatus. n = 4 independent biological repeats. *, p<0.025 and **, p<0.009 by two-tailed unpaired t-test.
c, Confocal fluorescence images of cells carrying activation apparatus (fused with fluorescent protein markers) and target gene. Sequential fluorescence images were acquired at distinct wavelengths for excitation laser and emission detector (433, 515, and 587 nm respectively for excitations, 440–475, 518–540, and 590–610 nm respectively for emission). Scale bar is 7.5 μm. n=3 independent biological repeats with similar results.
d, Normalized fluorescence signal profile over distance across the cell. Fluorescence signal of each channel was converted into gray scale and profiled across the cell.
e, Regulation of transcription performance through programming heterotypic interactions. Increasing the number of synthetic enhancer sites demonstrates significant increase of the RFP signal after 2 h of induction. n = 4 independent biological repeats. **, p<0.009 by two-tailed unpaired t-test. Bar graph represents mean± SD
